- Contributors
- Commercial Partners
- Announcements
- Bio-Formats
The solution for reading proprietary microscopy image data and metadata

What is Bio-Formats?
Bio-Formats is a software tool for reading and writing image data using standardized, open formats. Bio-Formats is a community driven project with a standardized application interface that supports open source analysis programs like ImageJ , CellProfiler and Icy , informatics solutions like OMERO and the JCB DataViewer , and commercial programs like MATLAB .
Bio-Formats documentation includes a complete version history, guides for using Bio-Formats as a plugin for ImageJ/Fiji, extensive information on which formats are supported and the level of that support, and also information for developers wanting to extend Bio-Formats or use it in their own software.
Read the Docs
Development and Licensing
Bio-Formats is developed by the Open Microscopy Environment consortium, including development teams at LOCI at the University of Wisconsin-Madison , University of Dundee and Glencoe Software . Licensing and citing information is on the OME licensing page .
File Formats
Bio-Formats 5 improves support for High Content Screening, time lapse imaging, digital pathology and other complex multidimensional image formats, reading over 150 file formats and their metadata to the OME data model standard.
Our Community
Bio-Formats is a community project and we welcome your input . You can find guidance on reporting a bug , upload files to our QA system for testing, and contact us via our mailing lists or forums. Further information about how the OME team works and how you can contribute to our projects is in the Contributing Developer Documentation .
Get started working with your images now
© 2005-2024 University of Dundee & Open Microscopy Environment. Creative Commons Attribution 4.0 International License
OME source code is available under the GNU General public license or more permissive open source licenses, or through commercial license from Glencoe Software Inc. OME, Bio-Formats, OMERO, IDR and their associated logos are trademarks of Glencoe Software Inc. , which holds these marks to protect them on behalf of the OME community.
[ Site Map ]
Version: 2024.06.28
Learn About Us
- What We Can Do
- What We’re Up To
Try Our Products
- OMERO Download
- Bio-Formats Download
- OME Files Download
Knowledge Base
- Documentation
- User Guides
- Security Advisories
Skip to content. | Skip to navigation
- Accessibility

| , and , informatics solutions like and the , and commercial programs like . Bio-Formats is developed by the Open Microscopy Environment consortium, including development teams at , and . Licensing and citing information is on the . Bio-Formats 5 improves support for High Content Screening, time lapse imaging, digital pathology and other complex multidimensional image formats, reading and converting over 140 to the data standard. Bio-Formats is a community project and we welcome your input. You can find guidance on , upload files to our for testing, and via our mailing lists or forums. Further information about how the OME team works and how you can contribute to our projects is in the . |
© 2000-2024 University of Dundee & Open Microscopy Environment. Creative Commons Attribution 4.0 International License
OME source code is available under the GNU General public license or through commercial license from Glencoe Software
ImageJ Docs
- User Guides
- Keyboard Shortcuts
- Tips and Tricks
- Troubleshooting
- Frequently Asked Questions
- Raspberry Pi
Bio-Formats
- Video formats
- Olympus VSI
- Principles of Scientific Imaging
- Annotating Images
- Colocalization
- Color Image Processing
- Deconvolution
- Image Intensity Processing
- Particle Analysis
- Registration
- Segmentation
- Stack-slice Manipulations
- T-functions
- Visualization
- Z-functions
- List of Extensions
- List of Update Sites
- Following an Update Site
- Creating an Update Site
- Terms of Service
- Automatic Upload
- Uploading to Core Sites
- Update Sites FAQ
- Batch Processing
- Scripting Basics
- Script Editor
- Using Javadoc
- Auto Import
- Running Headlessly
- Scripting Comparisons
- BeanShell Scripting
- Groovy Scripting
- ImageJ Macro
- Lisp (Clojure)
- Ruby (JRuby)
- Scala Scripting
- Architecture
Source code
- Project management
- Coding style
- IntelliJ IDEA
- Command Line
- Writing plugins
- Contributing to a plugin
- Development lifecycle
- Building a POM
- Hands-on debugging
- Adding new ops
- Adding new formats
- Using native libraries
- Tips for developers
- Tips for C++ developers
- ImageJ 1.x plugins
- Git in Eclipse (EGit)
- Git mini howto
- Git workshop
- Git conflicts
- Git topic branches
- Git reflogs
- Git submodules
- How to pinpoint regressions
- How to publish a git repository
- How to extract a subproject
- Contributors
- Organizations
- Contributing to Fiji
- Public domain
- Advanced Editing
- Source Code
- Math Expressions
- Reporting Issues
- Mailing Lists
- Presentations
- Conferences
- ImageJ Common
- ImageJ Legacy
- Getting Started
- Accessibles
- Introductory Workshop
- Advanced Workshop
- ImgLib2 images in MATLAB
- Developing ImgLib2
- ImgLib2 Discussion
Vital statistics
Loading... | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Source | none | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| License | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Release | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Date | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Development status | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Support status | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Page contents
Import data from many life sciences file formats, and export to several open formats. See LOCI’s Bio-Formats site for a historical narrative of the project. DocumentationWhat follows is a brief overview of the available plugins. You will find them all under the “Bio-Formats” submenu of Plugins. See the Bio-Formats website for additional information about Bio-Formats in general. See especially the Using Bio-Formats page for detailed instructions. Bio-Formats ImporterThe Bio-Formats Importer is a plugin for reading data into Fiji. It can open many dozens of proprietary life sciences formats, and standardize their acquisition metadata into a common OME data model . It will also extract and set basic metadata values such as spatial calibration if they are available in the file. Often, you will not need to worry about this plugin to import your data, because Bio-Formats is largely integrated with the File › Open command of Fiji. However, for certain file formats, you may wish to explicitly activate the Bio-Formats Importer to override the default behavior of Fiji. For example, by default Fiji uses some built-in logic to open TIFF files, but this logic may fail with certain TIFFs. The Bio-Formats Importer plugin may be able to import such TIFFs successfully. Bio-Formats ExporterThe Bio-Formats Exporter is a plugin for exporting data to disk. It can save to the open OME-TIFF file format, as well as several movie formats (e.g., QuickTime, AVI) and graphics formats (e.g., PNG, JPEG).
For OME-TIFF and TIFF formats, the file extensions *.tif and *.tiff are associated with the standard 32-bit TIFF, which has a 4GB size limit. On the other hand, the file extensions *.tf2 , *.tf8 , and *.btf are automatically associated with the 64-bit BigTIFF format , which can store much bigger data. Bio-Formats Remote ImporterThe Bio-Formats Remote Importer is a plugin for importing data from a remote URL. It is likely to be less robust than working with files on disk, so when possible we recommend downloading your data to disk and using the regular Bio-Formats Importer instead. Bio-Formats Windowless ImporterThe Bio-Formats Windowless Importer is a version of the Bio-Formats Importer plugin that runs with the last used settings to avoid popping up any additional dialogs beyond the file chooser. If you find that you always use the same import settings, you may wish to use the windowless importer to save time. Bio-Formats Macro ExtensionsThe Bio-Formats plugins come with a set of macro extensions to enable additional functionality from macros. The Bio-Formats Macro Extensions plugin prints out the available commands to the ImageJ log window, along with instructions for using them. Stack SlicerThe Stack Slicer plugin is a helper plugin used by the Bio-Formats Importer . It can also be used standalone to split a stack across channels, focal planes or time points. Bio-Formats Plugins ConfigurationThe Bio-Formats Plugins Configuration dialog is a useful way to configure the behavior of each file format. You can see a list of supported file formats on the Formats tab, toggle each format on or off (which is useful, for example, if your file is being detected as the wrong format), and toggle whether each format bypasses the importer options dialog using the “Windowless” checkbox. You can also configure any specific options for each format—for example, for QuickTime, you can toggle between Apple’s QTJava library or Bio-Formats’s built-in support. In addition, you can see a list of available helper libraries used by Bio-Formats on the Libraries tab. Bio-Formats Plugins Shortcut WindowThe Bio-Formats Plugins Shortcut Window is a small window with a quick-launch button for each plugin. You can also drag and drop files onto the shortcut window to open them quickly using the Bio-Formats Importer plugin. Update Bio-Formats PluginsThe Update Bio-Formats Plugins command will check online for updates to the Bio-Formats Plugins. In the case of Fiji, we recommend that you do not use this method of update, but instead use the ImageJ Updater . Calling Bio-Formats from the command lineYou can invoke Bio-Formats from the command line using the ImageJ Launcher :
Leave off the -batch flag if you want ImageJ to remain open afterward. Note that you cannot use the –headless option because Bio-Formats does not work in headless mode, even when running as a macro. (You will see VerifyError on the console if you try.) Here is an example macro created in such a fashion: Bio-Formats has a high-level scripting interface, accessible by Java and all scripting languages supported by Fiji (but not the ImageJ macro language). Java example: If needed, import options can be set: Daily buildsThe daily builds are not yet released and should be considered beta in quality. There may be new bugs. In particular, you should avoid exporting data using the Bio-Formats Exporter because the files it writes might not be readable later by release versions of Bio-Formats or other OME-compliant tools. Fiji ships release versions of Bio-Formats. However, given the long time frame between releases, you can update to the latest code by toggling the Bio-Formats update site, which includes the latest bug-fixes. When reporting a bug, the developers may ask you to test with the Bio-Formats update site enabled. To enable the Bio-Formats update site:
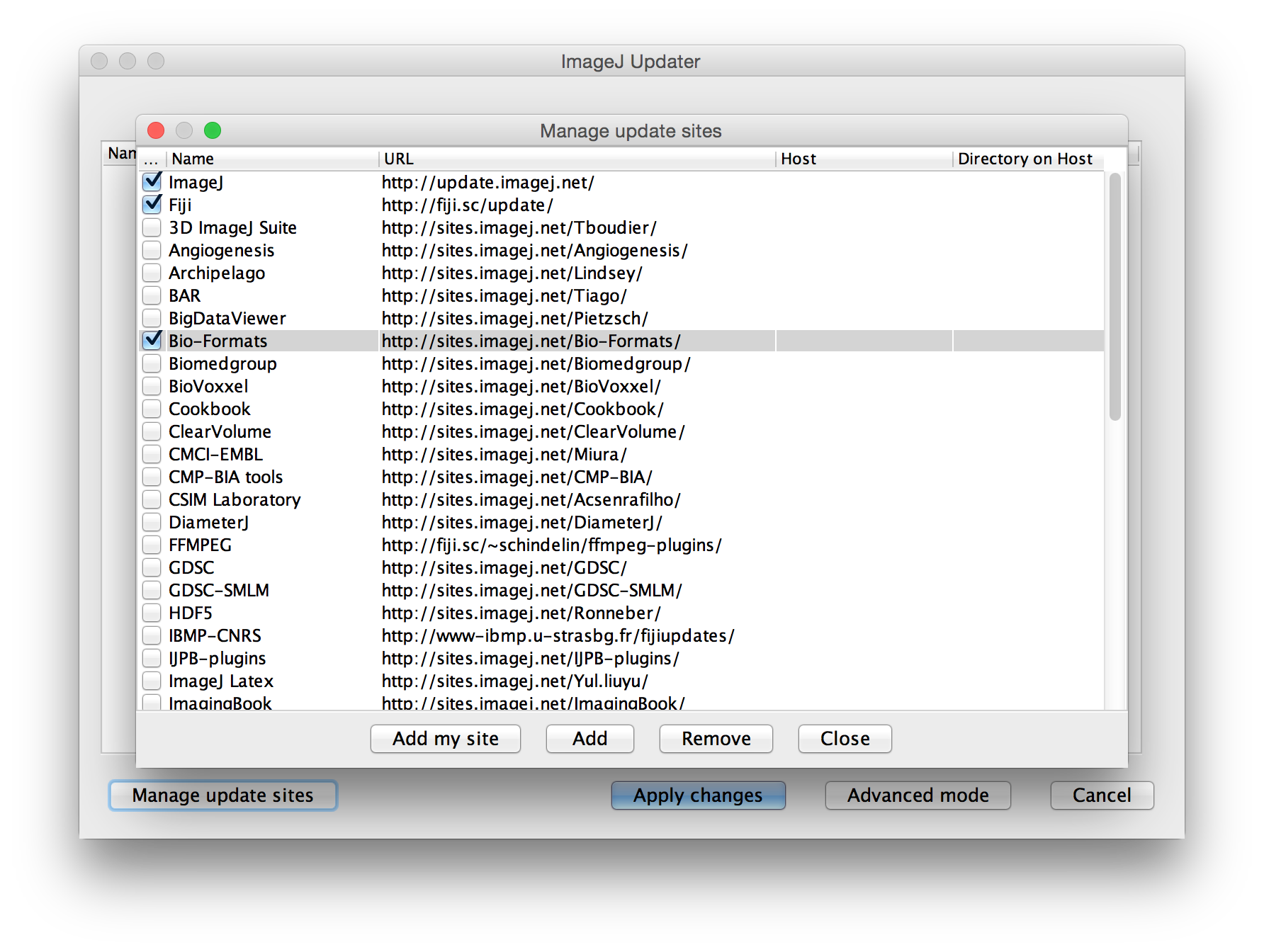
 The Bio-Formats source code is on GitHub . Reporting bugsTo report a bug in Bio-Formats, please see reporting a bug in Bio-Formats . Publicationdoi:10.1083/jcb.201004104
Home - Biofrontiers-ALMC/bioformats-matlab GitHub WikiWelcome to the Bioformats Image Toolbox wiki! The Bioformats Image Toolbox provides a way to read microscope files (such as Nikon's ND2 format) into MATLAB. The toolbox implements a MATLAB class around the Open Microscopy Environment group's Bio-Formats Image Package Java library. Example usageDownloads and installation. Please visit the Releases page to download the latest version. You only need the MATLAB toolbox ( .mltbx ) file, not the source code. Documentation
Bug report and Feature RequestsPlease submit bug reports or ideas for new features on the Issues Board . For bug reports, please provide enough details to reproduce the error, including details of external files used. If possible, please include a minimal example that produces the error and post that. Please also include the full error message at the end of the report. Known issues: 2020/04/13 - Working with ND2 files that have been cropped in Nikon Elements can result in odd problems such as images appearing out of order or timestamps being nonsensical. Workaround: If you are using Windows or Linux, you can try using an alternative ND2 reader that uses the official SDKs Contributer Guide
License and acknowledging the projectWe would appreciate an acknowledgement in your publications or presentations. An example acknowledgement statement is: This project uses the MATLAB Bioformats Image Toolbox developed by Dr. Jian Wei Tay of the BioFrontiers Advanced Light Microscopy Core at the University of Colorado Boulder. We also ask that you send us an email at [email protected] so we can keep track of usage stats. This helps us in reporting and securing funding for the facility. This project is provided under the MIT License. ⚠️ **GitHub.com Fallback** ⚠️

Bio-Formats
Page Contents
Previous topicQuick search. Enter search terms or a module, class or function name.
MATLAB is a high-level language and interactive environment that facilitates rapid development of algorithms for performing computationally intensive tasks. Calling Bio-Formats from MATLAB is fairly straightforward, since MATLAB has built-in interoperability with Java. We have created a set of scripts for reading image files. Note the minimum supported MATLAB version is R2007b (7.5). Installation ¶Download the MATLAB toolbox from the Bio-Formats downloads page . Unzip bfmatlab.zip and add the unzipped bfmatlab folder to your MATLAB path. As of Bio-Formats 5.0.0, this zip now contains the bundled jar and you no longer need to download loci_tools.jar or the new bioformats_package.jar separately. Please see Using Bio-Formats in MATLAB for usage instructions. If you intend to extend the existing .m files, please also see the developer page for more information on how to use Bio-Formats in general. Performance ¶In our tests (MATLAB R14 vs. java 1.6.0_20), the script executes at approximately half the speed of our showinf command line tool , due to overhead from copying arrays.  Upgrading ¶To use a newer version of Bio-Formats, overwrite the content of the bfmatlab folder with the newer version of the toolbox and restart MATLAB. Alternative scripts ¶Several other groups have developed their own MATLAB scripts that use Bio-Formats, including the following:
Navigation MenuSearch code, repositories, users, issues, pull requests..., provide feedback. We read every piece of feedback, and take your input very seriously. Saved searchesUse saved searches to filter your results more quickly. To see all available qualifiers, see our documentation .
A MATLAB class using the OME Bio-Formats library Biofrontiers-ALMC/bioformats-matlabFolders and files.
Repository files navigationBioformats image toolbox. Welcome to the Bioformats Image Toolbox! The Bioformats Image Toolbox provides a way to read microscope files (such as Nikon's ND2 format) into MATLAB. The toolbox implements a MATLAB class to use the Bio-Formats Image Package Java library. Prerequisites
Quick Navigation
This project uses the OME Bio-Formats Library . Acknowledging usWe would appreciate an acknowledgement in your publications or presentations. An example acknowledgement statement is: This project is developed by Dr. Jian Wei Tay of the BioFrontiers Advanced Light Microscopy Core at the University of Colorado Boulder. We also ask that you send us an email at [email protected] so we can keep track of usage stats. This helps us in reporting and securing funding for the facility. This project is provided under the MIT License.
You are now following this Submission
 Bioformats Image Toolbox
Welcome to the Bioformats Image Toolbox! The Bioformats Image Toolbox provides a way to read microscope files (such as Nikon's ND2 format) into MATLAB. The toolbox implements a MATLAB class to use the Bio-Formats Image Package Java library. Prerequisites
Quick Navigation
This project uses the OME Bio-Formats Library . Acknowledging usWe would appreciate an acknowledgement in your publications or presentations. An example acknowledgement statement is: This project is developed by Dr. Jian Wei Tay of the BioFrontiers Advanced Light Microscopy Core at the University of Colorado Boulder. We also ask that you send us an email at [email protected] so we can keep track of usage stats. This helps us in reporting and securing funding for the facility. This project is provided under the MIT License. JWTay (2024). Bioformats Image Toolbox (https://github.com/Biofrontiers-ALMC/bioformats-matlab/releases/tag/v1.2.3), GitHub. Retrieved July 5, 2024 . MATLAB Release CompatibilityPlatform compatibility, tags add tags, community treasure hunt. Find the treasures in MATLAB Central and discover how the community can help you! Discover Live EditorCreate scripts with code, output, and formatted text in a single executable document. Learn About Live Editor
Select a Web Site Choose a web site to get translated content where available and see local events and offers. Based on your location, we recommend that you select: . You can also select a web site from the following list How to Get Best Site Performance Select the China site (in Chinese or English) for best site performance. Other MathWorks country sites are not optimized for visits from your location.
Asia Pacific
Contact your local office Bioinformatics ToolboxRead, analyze, and visualize genomic and proteomic data. Have questions? Contact Sales . Bioinformatics Toolbox provides algorithms and apps for building bioinformatics pipelines, Next Generation Sequencing (NGS), microarray analysis, mass spectrometry, and gene ontology. You can read genomic and proteomic data and explore this data with the Genomics Viewer app and visualize with spatial heatmaps and clustergrams. The toolbox (with Statistics and Machine Learning Toolbox) provides statistical, regression, classification, and (with Deep Learning Toolbox) deep learning techniques to build complete analysis pipelines. The Biopipeline Designer app lets you interactively build bioinformatics pipelines for genomic data analysis, locally or in the cloud. You can use a combination of built-in bioinformatics pipeline blocks that integrate proven NGS libraries and custom blocks to extend analyses with community tools. You can create a pipeline to preprocess reads, map them to a reference genome, and perform statistical analysis, like RNA-Seq differential expression analysis or ChIP-Seq analysis.  Build Bioinformatics PipelinesWith the Biopipeline Designer app you can interactively build and run end-to-end bioinformatics pipelines, locally or in the cloud. Build a pipeline with built-in blocks that integrate proven NGS libraries or custom blocks to extend analyses with community tools for each step in the process. Run pipelines in parallel (using Parallel Computing Toolbox) and in batch mode. Documentation | Examples  Next Generation Sequencing (NGS)The toolbox provides algorithms and visualization techniques for NGS. For example, you can preprocess reads, map them to a reference genome, and perform statistical analyses, such as differential expression analysis from RNA-Seq data or ChIP-Seq data analysis. Documentation | Examples  Sequence AnalysisApply sequence analysis methods, including pairwise sequence, sequence profile, and multiple sequence alignment. Manipulate and evaluate sequences to gain a deeper understanding of your data. Perform BLAST searches against known sequences in online or local databases. Documentation | Example  Mass Spectrometry Data AnalysisBioinformatics Toolbox enables the analysis of SELDI, MALDI, LC/MS, and GC/MS data. You can smooth, align, and normalize spectra and use classification, statistical, and machine learning techniques to create classifiers and identify potential biomarkers.  Phylogenetic Tree AnalysisConstruct phylogenetic trees using hierarchical linkage with a variety of techniques, including neighbor joining, single and complete linkage, and Unweighted Pair Group Method Average (UPGMA).  Reading Genomic and Proteomic DataYou can read data from common file formats such as SAM, BAM, FASTA, FASTQ, GTF, and GFF and from online databases such as NCBI Gene Expression Omnibus, GenBank ® , and the Sequence Read Archive. You can use specialized data containers for data too large to fit in memory.  Statistical and Machine Learning AlgorithmsBioinformatics Toolbox provides functions that build on the Statistics and Machine Learning Toolbox , which offers interactive tools for feature selection, classification, regression, mapping, and displaying hierarchy plots and pathways.  Microarray Data AnalysisNormalize microarray data using a variety of methods. Identify differentially expressed genes and perform enrichment analysis of expression results using gene ontology. Visualize gene and protein-protein interaction networks using graph theory algorithms. Deploying and Sharing AppsTurn your data analysis program into a customized software application. Build custom user interfaces; integrate with existing C, C++, and Java™ applications; and deploy standalone apps. Product Resources: "MATLAB enables young biologists to learn enough programming and math without being afraid of the code. They can write in MATLAB as if it were English." Dr. Jonas Almeida, Medical University of South Carolina Get a Free Trial30 days of exploration at your fingertips. Ready to Buy?Get pricing information and explore related products. Are You a Student?Your school may already provide access to MATLAB, Simulink, and add-on products through a campus-wide license. What's Next?CUSTOMER STORY Centers for Disease Control and Prevention Automates Poliovirus Sequencing and TrackingSequence viewer app. SUPPORT PACKAGE Bioinformatics Toolbox Software Support PackagesSelect a Web Site Choose a web site to get translated content where available and see local events and offers. Based on your location, we recommend that you select: . You can also select a web site from the following list How to Get Best Site Performance Select the China site (in Chinese or English) for best site performance. Other MathWorks country sites are not optimized for visits from your location.
Asia Pacific
Contact your local office Matlab Toolbox not containing latest version of bioformats_package.jar?I downloaded the Matlab Toolbox from Bio-Formats Downloads (OME) , but when I ran bfUpgradeCheck I receive the following: A new stable version of Bio-Formats is available version = char(loci.formats.FormatTools.VERSION); Returns version=5.1.2 rather than the expected 6.6.1 or equivalent. Should Matlab’s jar be behind the current release? And if not, can the 6.6.1 Bio-Formats Package be switched out in the Matlab files? I checked the newly-downloaded version because I was receiving a “Java Exception Error: Missing Library Java Advanced Imaging (JAI)”. I tried to switch out bioformas_package.jar(6.6.1) and then received: log4j:WARN No appenders could be found for logger (loci.formats.ClassList). log4j:WARN Please initialize the log4j system properly. ERROR: Java exception occurred: loci.formats.FormatException: No IFDs found I am running MATLAB 2021a and have Java 8 SE. Any help greatly appreciated. Hi @KE06 , I double checked the latest version of the toolbox and the jar it contains is the correct 6.6.1 version. What looks to be happening is a conflict with a previous version which has been installed. If you run the command javaclasspath do you see any other versions of bioformas_package in a different location? Hi @dgault my dynamic path returns a single instance of bioformats_package.jar in my current working folder. Static path returns the expected series. As reference: I downloaded the Matlab Toolbox from the website, deleted my prior bioformats folder, and pasted in the freshly downloaded folder. Within Matlab if you go to your current working folder, expand the bioformats_package.jar and look for the file META-INF/MANIFEST.MF and open it, you should see details as below. Does your jar show the version as 6.6.1 here? Thanks @dgault for your previous assistance! From your help and for future readers, I was able to determine that running my prior code was calling a previous instance of Bioformats elsewhere within my computer (v 5.1.2) regardless of having v6.6.1 packaged in the same folder as my Matlab script. I admit to not knowing why this happened (or specifically where in the computer it was being called from) but changing the computer path of the image I was calling resolved: Java Exception Error: Missing Library Java Advanced Imaging (JAI) Furthermore, Turned out to be the original image file was also corrupted and could not be viewed by our Whole Slide Image viewer independent of Bioformats. Related Topics
Collectives™ on Stack OverflowFind centralized, trusted content and collaborate around the technologies you use most. Q&A for work Connect and share knowledge within a single location that is structured and easy to search. Get early access and see previews of new features. BioFormats In MatlabI am attempting to implement bio-formats into matlab to run with matlab code the process nd2 microscopy images and quantify them. I am trying to within Matlab directly convert files from nd2>tiff format. If anyone has any guidance as to how I can do this, Please let me know. I have tried using the bfconvert function and it doesn't seem to work. I have also downloaded bftools.zip and bfmatlab.zip.I am wondering if I have to do an integration to allow matlab code to properly function with Java language.
Know someone who can answer? Share a link to this question via email , Twitter , or Facebook .Your answer. Reminder: Answers generated by artificial intelligence tools are not allowed on Stack Overflow. Learn more Sign up or log inPost as a guest. Required, but never shown By clicking “Post Your Answer”, you agree to our terms of service and acknowledge you have read our privacy policy . Browse other questions tagged image matlab image-processing file-conversion or ask your own question .
Hot Network Questions
 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
IMAGES
VIDEO
COMMENTS
MATLAB¶. MATLAB is a high-level language and interactive environment that facilitates rapid development of algorithms for performing computationally intensive tasks.. Calling Bio-Formats from MATLAB is fairly straightforward, since MATLAB has built-in interoperability with Java. We have created a toolbox for reading and writing image files. Note the minimum recommended MATLAB version is R2017b.
Bio-Formats Package bioformats_package.jar. ... A bundle of functions for using Bio-Formats from MATLAB with bioformats_package.jar already included. For more info, see the MATLAB documentation. Download (29.97 MB) Octave Package bioformats-octave-7.3..tar.gz.
MATLAB¶. MATLAB is a high-level language and interactive environment that facilitates rapid development of algorithms for performing computationally intensive tasks.. Calling Bio-Formats from MATLAB is fairly straightforward, since MATLAB has built-in interoperability with Java. We have created a set of scripts for reading image files. Note the minimum supported MATLAB version is R2007b (7.5).
Note the minimum supported MATLAB version is R2007b (7.5). As described in Using Java Libraries, every installation of MATLAB includes a JVM allowing use of the Java API and third-party Java libraries. All the helper functions included in the MATLAB toolbox make use of the Bio-Formats Java API. Please refer to the Javadocs for more information.
The Bioformats Image Toolbox provides a way to read microscope files (such as Nikon's ND2 format) into MATLAB. The toolbox implements a MATLAB class to use the Bio-Formats Image Package Java library. Prerequisites. MATLAB R2016b or newer (it is highly likely that an earlier version will work).
Bio-Formats is a software tool for reading and writing image data using standardized, open formats. Bio-Formats is a community driven project with a standardized application interface that supports open source analysis programs like ImageJ, CellProfiler and Icy, informatics solutions like OMERO and the JCB DataViewer, and commercial programs like MATLAB.
Help installing Bio-Formats. Learn more about bio-format, image processing, image
Changes: [NEW] Added new properties to read globalMetadata; Usage: To install, download the toolbox file (.mltbx) and open the file in MATLAB. For more details, please see the wiki
Bio-Formats is a software tool for reading and writing image data using standardized, open formats. Bio-Formats is a community driven project with a standardized application interface that supports open source analysis programs like ImageJ, CellProfiler and Icy, informatics solutions like OMERO and the JCB DataViewer, and commercial programs like Matlab.
Bio-Formats Importer. The Bio-Formats Importer is a plugin for reading data into Fiji. It can open many dozens of proprietary life sciences formats, and standardize their acquisition metadata into a common OME data model. It will also extract and set basic metadata values such as spatial calibration if they are available in the file.
The toolbox implements a MATLAB class around the Open Microscopy Environment group's Bio-Formats Image Package Java library. Example usage % Create a new BioformatsImage object bfr ... This project uses the MATLAB Bioformats Image Toolbox developed by Dr. Jian Wei Tay of the BioFrontiers Advanced Light Microscopy Core at the University of ...
It is using the MATLAB wrapper for BioFormats to read CZI files (and other 3rd party image formats as well) including some of the relevant meta-information. Use it at your own risk … image 1323×623 96.6 KB
MATLAB¶. MATLAB is a high-level language and interactive environment that facilitates rapid development of algorithms for performing computationally intensive tasks.. Calling Bio-Formats from MATLAB is fairly straightforward, since MATLAB has built-in interoperability with Java. We have created a set of scripts for reading image files. Note the minimum supported MATLAB version is R2007b (7.5).
Filenamein = Dirlist (Filecounter).name. Newimagename = fullfile (Directory, Filenamein); Newimage = bfopen (Newimagename); OR use your existing code except. Theme. Copy. Newimage = bfopen (char (Newimagename)); bfopen () does not handle string objects. Your Directory is a string object and you used "+" to concatenate to it, producing a string ...
MATLAB¶. MATLAB is a high-level language and interactive environment that facilitates rapid development of algorithms for performing computationally intensive tasks.. Calling Bio-Formats from MATLAB is fairly straightforward, since MATLAB has built-in interoperability with Java. We have created a set of scripts for reading image files. Note the minimum supported MATLAB version is R2007b (7.5).
Bio-Formats 6.0.0 bumps the minimal supported Java version to 8 [1]. For MATLAB users, this means it is no longer possible to use old versions of MATLAB shipping JVM 7 natively with Bio-Formats 6. There are two possibilities: if possible, upgrading to MATLAB R2017b or later is recommended.
For reference, all of my work/files are in '\Users\user_name\Documents\MATLAB\Image Analysis' I pasted all of the files from the Bio-Formats website directly in this 'Image Analysis' folder, along with the rest of my .m files. Yet I can't use Bio-Formats.
Bioformats Image Toolbox. Welcome to the Bioformats Image Toolbox! The Bioformats Image Toolbox provides a way to read microscope files (such as Nikon's ND2 format) into MATLAB. The toolbox implements a MATLAB class to use the Bio-Formats Image Package Java library.
Download and share free MATLAB code, including functions, models, apps, support packages and toolboxes
Bioinformatics Toolbox provides algorithms and apps for building bioinformatics pipelines, Next Generation Sequencing (NGS), microarray analysis, mass spectrometry, and gene ontology. You can read genomic and proteomic data and explore this data with the Genomics Viewer app and visualize with spatial heatmaps and clustergrams.
Hi @dgault my dynamic path returns a single instance of bioformats_package.jar in my current working folder. Static path returns the expected series. As reference: I downloaded the Matlab Toolbox from the website, deleted my prior bioformats folder, and pasted in the freshly downloaded folder.
I am attempting to implement bio-formats into matlab to run with matlab code the process nd2 microscopy images and quantify them. I am trying to within Matlab directly convert files from nd2>tiff format. If anyone has any guidance as to how I can do this, Please let me know. I have tried using the bfconvert function and it doesn't seem to work.