
An official website of the United States government, Department of Justice.
Here's how you know
Official websites use .gov A .gov website belongs to an official government organization in the United States.
Secure .gov websites use HTTPS A lock ( Lock A locked padlock ) or https:// means you’ve safely connected to the .gov website. Share sensitive information only on official, secure websites.

Solving Cold Cases with DNA: The Boston Strangler Case

This was a ghastly crime.
Nineteen-year-old Mary Sullivan had just moved from Cape Cod to Boston, where she rented an apartment in the bustling Beacon Hill neighborhood. Within a few days of her arrival in January 1964, she was found dead. Her attacker raped her and strangled her to death.
Sullivan was one of 11 women whom Albert DeSalvo — known as the Boston Strangler — would later confess to killing. However, he then recanted, leaving lingering doubts about the possibility that the real assailant had eluded capture.
DeSalvo was never convicted of any of the Strangler killings, but he was sentenced to life in prison on other rape charges. He was stabbed to death in 1973. For decades after his death, experts argued about whether he really was the Strangler or whether someone else committed the crimes and got away.
Evidence that finally linked DeSalvo to the Sullivan assault emerged in July 2013.
DNA Provides Answers
Over the years, NIJ has funded the examination of "cold cases" across the country through its Solving Cold Cases with DNA program. The funding helps police departments identify, review, investigate and analyze violent crime cold cases that could be solved through DNA analysis. Sometimes the cases are so old that DNA testing did not yet exist when the crimes were committed, and testing biological evidence now might show a match with a suspect.
In 2009 and 2012, the city of Boston received competitive grants under NIJ's cold case program. The Boston Police Department's cold case squad decided to use some of the NIJ funding to test DNA from a nephew of DeSalvo's and look for a match with seminal fluid that had been found on Sullivan's body and on a blanket at the crime scene. When forensics experts ran the test, they got a hit.
The match was possible because of tests that zero in on short tandem repeats (STRs), which are patterns found on DNA strands. Forensic scientists use a specialized test that focuses on male (Y) chromosomes. Y-chromosome DNA comes from fathers who pass their Y-STR DNA profiles to their male offspring. Barring a mutation, the profiles remain unchanged. Every male in a paternal lineage has the same Y-STR DNA profile. This includes fathers, sons, brothers, uncles, nephews and a wider group of male relatives, even out to third and fourth cousins.
NIJ has funded research on Y-STRs for years, believing that it would give forensics experts a powerful and important tool in certain cases.
Testing of Y-STRs in the Mary Sullivan case showed a match between DNA from the crime scene and DeSalvo's nephew. According to Boston officials, this match implicated DeSalvo and excluded 99.9 percent of the male population. But because a Y-STR profile is common to a group of male family members, it does not yield the more precise match to a particular individual available in other DNA tests.
Armed with the Y-STR testing results, Boston authorities went a step further and exhumed DeSalvo's body in July 2013 so they could conduct a confirmatory test using a DNA sample directly from DeSalvo. DNA extracted from a femur and three teeth yielded a match — specifically, DNA specialists calculated the odds that a white male other than DeSalvo contributed the crime scene evidence at one in 220 billion — leaving no doubt that DeSalvo had raped and murdered Mary Sullivan.
NIJ's Solving Cold Cases with DNA Program
Since 2005, NIJ has awarded more than $73 million to more than 100 state and local law enforcement agencies through its Solving Cold Cases with DNA competitive grant program. This funding has allowed the agencies to review more than 119,000 cases. The funding has also facilitated the entry of almost 4,000 DNA profiles into the FBI's Combined DNA Index System, yielding more than 1,400 hits.
The program has given agencies the opportunity to put resources toward solving homicides, sexual assaults and other violent offenses that otherwise might never have been reviewed or reinvestigated. Crime scene samples from these cases — previously thought to be unsuitable for testing — have yielded DNA profiles. And samples that previously generated inconclusive DNA results have been reanalyzed using modern technology and methods.
Thanks to these cold case funds and the latest Y-STR technology, the Boston Police Department was able to solve the mystery surrounding Mary Sullivan almost 50 years after her death.
For More Information
- Read more about the Solving Cold Cases with DNA program.
- To learn more about STR analysis, read "STR Analysis" from issue 267 of the NIJ Journal .
About This Article
This article appeared in NIJ Journal Issue 273 , March 2014.
About the author
Philip Bulman is a former NIJ writer and editor.
Cite this Article
Read more about:, related publications.
- NIJ Journal Issue No. 273
- Skip to main content
- Keyboard shortcuts for audio player
Detectives Just Used DNA To Solve A 1956 Double Homicide. They May Have Made History
Sharon Pruitt-Young

Clippings from the Great Falls Tribune were part of the Cascade County Sheriff's Office investigative file into the 1956 murders of Patricia Kalitzke and Lloyd Duane Bogle. Traci Rosenbaum/USA Today Network via Reuters Co. hide caption
Clippings from the Great Falls Tribune were part of the Cascade County Sheriff's Office investigative file into the 1956 murders of Patricia Kalitzke and Lloyd Duane Bogle.
It was only three days into 1956 when three boys from Montana, out for a hike on a normal January day, made a gruesome discovery they were unlikely to ever forget.
During a walk near the Sun River, they found 18-year-old Lloyd Duane Bogle, dead from a gunshot wound to the head. They found him on the ground near his car, and someone had used his belt to tie his hands behind his back, according to a report from the Great Falls Tribune . The next day brought another disturbing discovery: A county road worker found 16-year-old Patricia Kalitzke's body in an area north of Great Falls, the paper reports. She had been shot in the head, just as Bogle had been, but she had also been sexually assaulted.
Their killings went unsolved until this week when investigators announced they had cracked what is believed to be the oldest case solved with DNA and forensic genealogy.
The victims were discovered in a lover's lane
Bogle, an airman hailing from Texas, and Kalitzke, a junior at Great Falls High School, had fallen for each other and were even considering marriage, the Tribune reports. The place where they were believed to have been killed was a known "lover's lane," according to a clipping from a local newspaper posted on a memorial page.
But their love story was brutally cut short by the actions of a killer whose identity would not be revealed for more than 60 years. And it was not for lack of trying: Early on in the case, investigators followed numerous leads, but none of them panned out. The case eventually went cold.
For decades, the Cascade County Sheriff's Office continued to work on it, with multiple detectives attempting to make progress over the years. One such investigator was Detective Sgt. Jon Kadner, who was assigned the case in 2012 — his first cold case, he said during an interview with NPR. He was immediately met with the daunting task of digitizing the expansive case file, an endeavor that took months.
He continued to work on the Kalitzke/Bogle case even while handling the newer cases that were landing on his desk all the time, but he had a feeling that more was needed to get to the bottom of what had happened to the couple all those decades ago.
"My first impression was that the only way we're gonna ever solve this is through the use of DNA," Kadner said.
Detectives turned to a new forensic investigation
Fortunately, Kadner had something to work with. During Kalitzke's autopsy in 1956, coroners had taken a vaginal swab, which had been preserved on a microscopic slide in the years since, according to the Great Falls Tribune report. Phil Matteson, a now-retired detective with the sheriff's office, sent that sample to a local lab for testing in 2001, and the team there identified sperm that did not belong to Bogle, her boyfriend, the paper reports.
Armed with this knowledge, Kadner in 2019 sought out the assistance of Bode Technology. After forensic genealogy was used to finally nab the Golden State Killer the year prior, law enforcement officials were becoming increasingly aware of the potential to use that technology to solve cold cases — even decades-old cases like Kalitzke and Bogle's.
With the help of partnering labs, forensic genealogists are able to use preserved samples to create a DNA profile of the culprit and then use that profile to search public databases for any potential matches. In most cases, those profiles can end up linking to distant relatives of the culprit — say, a second or third cousin. By searching public records (such as death certificates and newspaper clippings), forensic genealogists are then able to construct a family tree that can point them right to the suspect, even if that suspect has never provided their DNA to any public database.
In this case, "Our genealogists, what they're going to do is independently build a family tree from this cousin's profile," Andrew Singer, an executive with Bode Technology, told NPR. He called it "a reverse family tree. ... We're essentially going backwards. We're starting with a distant relative and trying to work back toward our unknown sample."
It worked: DNA testing led investigators to a man named Kenneth Gould. Before moving to Missouri in 1967, Gould had lived with his wife and children in the Great Falls area around the time of the murders, according to the Tribune .
"It felt great because for the first time in 65 years we finally had a direction and a place to take the investigation," Kadner told NPR. "Because it was all theories up to that point ... we finally had a match and we had a name. That changed the whole dynamic of the case."
Investigators' goal is a safer world
But there was one big problem: Gould had died in 2007 and his remains had been cremated, according to the Tribune . The only way to prove his guilt or his innocence was to test the DNA of his remaining relatives.
Detectives had an uncomfortable task ahead of them: letting a dead man's family know that, despite the fact that he'd never previously been identified as a person of interest, he was now the key suspect in a double homicide and rape.
Authorities traveled to Missouri, where they spoke with Gould's children and told them about the Kalitzke/Bogle case and eventually identified their father as a suspect, Kadner said. They asked for the family's help in either proving or disproving that Gould was the man responsible and the family complied.
The test results said Gould was the guy. With the killer finally identified, Kadner was able to reach out to the victims' surviving relatives and deliver the closure that had taken more than 60 years to procure. It was a bittersweet revelation: They were grateful for answers, but for many of the older people in the family, it was a struggle to have those wounds reopened.
"They're excited, but at the same time, it has brought up a lot of memories," Kadner said.
Now, the sheriff's office is considering forming a cold case task force, as other law enforcement agencies have done. The hope is that they'll be able to provide more families with the answers they deserve and, in many cases, have spent years waiting for.
"If there's new technology and we are able to potentially solve something, we want to keep working at it, because ultimately we're trying to do it for the family," he said. "Give them some closure."
The Kalitzke/Bogle case is one of the oldest criminal cases that has been solved using forensic genealogy, and authorities are hopeful that they'll be able to use this ever-advancing technology to solve cold cases dating back even further — although new state legislation restricting forensic genealogy could complicate matters.
Even without that complication, Singer explained to NPR, the success rate depends heavily on how well the evidence has been preserved over the years. Still, he hopes that it can be used to help law enforcement improve public safety and "[prevent] tomorrow's victim."
"It's really fantastic technology and it's going to solve a lot of cold cases," Singer said.
How Forensic DNA Evidence Can Lead to Wrongful Convictions
Forensic DNA evidence has been a game-changer for law enforcement, but research shows it can contribute to miscarriages of justice.

Lynette White was murdered in 1988 . When the three men first imprisoned for her murder were found to have been wrongfully convicted, it seemed that her killer would go unpunished. However, new technology invented in 2002 was used to analyze DNA found at the scene of the murder. The only match was to a boy too young to have committed the murder, but DNA samples were taken from his family. The youth’s uncle confessed, and was sentenced to life imprisonment in 2003.

In criminal investigation, DNA evidence can be a game-changer. But DNA is just one piece of the puzzle, rarely giving a clear “he did it” answer. According to a consortium of forensic experts who released a report earlier this year, there are limits to what DNA can tell us about a crime. And what it can and can’t reliably prove in court needs to be much clearer.
Audio brought to you by curio.io
DNA (deoxyribonucleic acid) is a code that programs how we will develop, grow, and function. Humans are thought to have DNA that is 99.9% identical, but the remaining 0.1% makes us individuals, marking us out as unique. The fact that humans and chimpanzees have just a 1% difference in their DNA further highlights how meaningful a small difference can be. Generally, the more closely related we are to someone, the more similar our DNA will be to theirs.
The tiny part of our DNA that is unique to us can be used to generate a DNA profile. This profile is usually represented as a graph showing different peaks, which reports the patterns at different points where our DNA is most likely to be unique.
“The increasingly prominent role played by forensic science in the administration of criminal justice is due in no small measure to the meteoric rise in DNA profiling,” wrote the law professor Liz Hefferman in a 2008 article for the British Journal of Criminology .
DNA profiling has had some remarkable successes, including finally ending a two-decade long hunt for the “Green River Killer,” who strangled at least fifty women, dumping their bodies in various spots around the Green River in Washington State. However, DNA profiles are often not clean enough to conclusively identify an individual. Ideally, a DNA sample would be complete enough to examine at least 16 different “markers,” points at which an individual’s DNA fingerprint can be sketched out. But when DNA is damaged, as it often is through exposure to moisture or extreme temperatures, only some of these markers will be available, and forensics teams will generate a partial profile. Put simply, if a DNA profile is a complete description of a person’s appearance, a partial profile might describe only one of their traits—hair color, for instance.
Partial profiles will match up with many more people than a full profile. And even full profiles may match with a person other than the culprit. Further complicating matters, a single DNA profile might be mistakenly generated when samples from multiple people are accidentally combined. It’s a messy world.
Realistically, then, DNA profiles should only be thought of as being likely to have come from a specific individual. Statistical approaches such as “match probability,” which is based on comparisons between crime scene DNA and a hypothetical “random” person, often are misunderstood. A more rigorous statistical approach is likelihood ratio, which directly compares two hypotheses: the likelihood of the DNA coming from the suspect vs. the likelihood of the DNA coming from someone else. If the likelihood ratio is less than one, the defense position (the DNA is not the suspect’s) is better supported; if it is greater than one, there is more support for the prosecution case. Still, the ratio at most provides scientific support for a theory, not a yes-or-no answer.
A study from the University of California published in Law and Human Behavior tested undergraduate students’ abilities to interpret statistical evidence as it would be presented in court by prosecution and defense attorneys. The researchers found that the majority of these undergraduates failed to detect errors in statistical arguments and “made judgements based on fallacious reasoning.”
When the American Bar Association reported on DNA technology, it backed the use of DNA evidence, but urged caution in how statistics were interpreted. The ABA urged lawyers not to oversell DNA evidence and suggested that courts take the standards of the lab into account when considering DNA evidence. “Telling a jury it is implausible that anyone besides the suspect would have the same DNA test results is seldom, if ever, justified,” the report states.
In addition, the European Forensic Genetics Network of Excellence ( EUROFORGEN ) and the charity Sense about Science collaborated on a report released earlier this year . The report sought to clarify what DNA analysis can and cannot do within the criminal justice system. EUROFORGEN researcher Denise Sydercombe Court, based at King’s College London, said:
We all enjoy a good crime drama and although we understand the difference between fiction and reality, the distinction can often be blurred by overdramatised press reports of real cases. As a result, most people have unrealistic perceptions of the meaning of scientific evidence, especially when it comes to DNA, which can lead to miscarriages of justice.
At times, DNA evidence has been misused or misunderstood, leading to miscarriages of justice. A man with Parkinson’s disease who was unable to walk more than a few feet without assistance was convicted of a burglary based on a partial DNA profile match. His lawyer insisted on more DNA tests, which exonerated him. In 2011, Adam Scott’s DNA matched with a sperm sample taken from a rape victim in Manchester —a city Scott, who lived more than 200 miles away, had never visited. Non-DNA evidence subsequently cleared Scott. The mixup was due to a careless mistake in the lab, in which a plate used to analyze Scott’s DNA from a minor incident was accidentally reused in the rape case.
Then there’s the uncomfortable and inconvenient truth that any of us could have DNA present at a crime scene—even if we were never there. Moreover, DNA recovered at a crime scene could have been deposited there at a time other than when the crime took place. Someone could have visited beforehand or stumbled upon the scene afterward. Alternatively, their DNA could have arrived via a process called secondary transfer, where their DNA was transferred to someone else, who carried it to the scene.
Additionally, DNA technology is becoming more and more sensitive, but this is a double-edged sword. On one hand, usable DNA evidence is more likely to be detected than ever before. On the other hand, contamination DNA and DNA that arrived by secondary transfer is now more likely to be detected, confusing investigations. If legal and judicial personnel aren’t fully trained in how to interpret forensic and DNA evidence, it can result in false leads and miscarriages of justice.
Another consideration is that people shed DNA at different rates. DNA is found in bodily fluids, such as blood, semen, and saliva, but we also lose microscopic pieces of skin and hair on a regular basis. Some people lose DNA more quickly than others—if they have a skin condition, for example. If a thief uses a particular location as a stash, and a caretaker who suffers from eczema stumbles on it and reports it to the police, the forensics alone might implicate the caretaker. The quantity of their DNA present might suggest a significant period of time spent at that place. But in fact, the caretaker’s eczema resulted in more DNA being deposited there over a shorter time period.
Once a Week
Get your fix of JSTOR Daily’s best stories in your inbox each Thursday.
Privacy Policy Contact Us You may unsubscribe at any time by clicking on the provided link on any marketing message.
National DNA databases, then, present some ethical quandaries. Many cases would never have been solved if not for DNA databases. In the Lynette White case, the breakthrough came when the police obtained the DNA profile of a relative of the murderer. However, the retention of DNA details raises legitimate privacy concerns, especially in the context of familial searching. Partial matches are more likely to lead to false positive identification of suspects who are already in the DNA database. Given that less privileged groups tend to be over-represented in DNA databases, this is a serious issue.
In 2011, a group of scientists asked whether forensic DNA databases increase racial disparities in policing . They pointed out that, in the U.S., different communities are differently policed, leading to different rates of incarceration and DNA recording. According to the study authors, actual drug use is relatively higher in white communities, but “buy and bust” operations by police are more common in African American and Latino communities, leading to disproportionate arrests.
The lesson of all this research: DNA evidence is a powerful tool in criminal investigation and prosecution, but it must be used with care. It should never be oversold in court, and it should only ever be considered in light of other available evidence. For example, if DNA is recovered in a kitchen that has been broken into, it could be from the homeowner, their guests, or even a member of the CSI team (if sufficient care hasn’t been taken to avoid contamination). If a tool-mark impression reveals that a screwdriver was used to force open the window, and DNA is recovered from a screwdriver found at the scene that does not belong to the homeowner, that’s incriminating. If that DNA is a partial or full match with an individual with the same shoe size as a footprint left in the grass under the window, even more so. If that individual has a torn piece of clothing that matches cloth fibres snagged in the window, that’s more incriminating still. If digital evidence such as their mobile phone records place them at the scene at the time the break-in happened—even though they claim to have been elsewhere—then you have a more complete picture.
Editor’s notes: An earlier version of this story contained an unclear reference to evidence seized by police investigating the murder of Meredith Kercher. The example has since been removed. We regret any error.
A study cited in an earlier version of this article is no longer available for free on JSTOR .

JSTOR is a digital library for scholars, researchers, and students. JSTOR Daily readers can access the original research behind our articles for free on JSTOR.
Get Our Newsletter
More stories.
- The Dangers of Animal Experimentation—for Doctors

Witnessing and Professing Climate Professionals

Crucial Building Blocks of Life on Earth Can More Easily Form in Outer Space

Making Implicit Racism
Recent posts.
- Zheng He, the Great Eunuch Admiral
- Fredric Wertham, Cartoon Villain
- Separated by a Common Language in Singapore
- Katherine Mansfield and Anton Chekhov
Support JSTOR Daily
Sign up for our weekly newsletter.

An official website of the United States government, Department of Justice.
Here's how you know
Official websites use .gov A .gov website belongs to an official government organization in the United States.
Secure .gov websites use HTTPS A lock ( Lock A locked padlock ) or https:// means you’ve safely connected to the .gov website. Share sensitive information only on official, secure websites.
Solving Cold Cases with DNA: The Boston Strangler Case
This article explains how the National Institute of Justice's (NIJ's) funding under its Solving Cold Cases with DNA program ("DNA program"), specifically its funded research on Y-STRs, was the key to solving the mystery of Mary Sullivan's rape and murder in Boston almost 50 years after her death.
Although Albert DeSalvo, the purported "Boston Strangler," was suspected in Sullivan's rape and murder in January 1964, the case remained unsolved until after DeSalvo's death in prison in 1973. In 2009 and 2012, the city of Boston received competitive grants under NIJ's DNA program. The Boston Police Department's cold case squad decided to use some of the NIJ funding to test DNA from a nephew of DeSalvo's in order to seek a match with seminal fluid that had been found on Sullivan's body and a blanket at the crime scene; they got a match. The match was possible because of years of NIJ DNA funding of research on Y-STRs. Y-chromosome DNA profiles come from fathers who pass their Y-STR DNA profiles to their male offspring. Barring a mutation, the profiles remain unchanged. Every male in a paternal lineage has the same Y-STR DNA profile. This includes fathers, sons, brothers, uncles, nephews, and a wider group of male relatives, even to third and fourth cousins. Boston authorities went a step further and exhumed DeSalvo's body in July 2013, so they could conduct a confirmatory test using a DNA sample directly from DeSalvo. DNA extracted from a femur and three teeth yielded a match, leaving no doubt that DeSalvo had raped and murdered Mary Sullivan.
Additional Details
Related topics, similar publications.
- The Over-citation of Daubert in Forensic Anthropology
- A Conserved Interdomain Microbial Network Underpins Cadaver Decomposition Despite Environmental Variables
- Overcoming Recruiting Shortages by Applying Industrial and Organizational Psychology Practices
A Simplified Guide To
Dna evidence, how it’s done, sources of dna evidence.
The biological material used to determine a DNA profile include blood, semen, saliva, urine, feces, hair, teeth, bone, tissue and cells.
Samples that May be Used
Investigators collect items that could have been touched or worn by persons involved in a crime. The following items may contain DNA material:
- Sexual assault evidence kits
- Underclothes
- Dirty laundry
- Fingernail scrapings
- Cups/bottles
- Facial tissue
- Ligatures (rope, wire, cords)
- Stamps or envelopes
The best evidence occurs when a person’s DNA is found where it is not supposed to be. For example, consider a breaking-and-entering that occurred in a residential area. Near the point of forced entry, a knit cap was found which the homeowners confirm was not theirs. Several head hairs were recovered from the inside, one of which had a root with tissue attached, which made it possible to obtain a DNA profile. The DNA profile was used to identify the perpetrator.

A crime scene investigator uses a swab to collect blood from a crime scene. (Courtesy of NFSTC)

A cigarette butt found at a crime scene may contain valuable DNA material in the dried saliva. (Courtesy of NFSTC)

DNA evidence from both the victim’s blood and the perpetrator’s skin cells may be available from this hammer. (Courtesy of NFSTC)
As technology advances, forensic scientists are able to analyze smaller and smaller biological samples to develop a DNA profile. For example, if a person touched an object or weapon, skin cells may have been left behind. This low-level DNA is sometimes referred to as “touch DNA”. It can even be collected from a victim’s skin or bruises where they were handled roughly. Low-level DNA samples may be helpful when examining evidence where it would be difficult to retrieve fingerprints—such as textured surfaces on gun handles or automobile dashboards. However, not all jurisdictions have the capability to process this evidence.
To compare the victim’s or suspect’s DNA profile to the recovered crime-scene DNA, the laboratory will need to have their known biological samples available for a side-by-side comparison. These known samples are called reference samples . In some jurisdictions, a DNA sample is routinely taken from an arrestee during the process of booking and fingerprinting. However, this is an evolving area of law and states vary in their laws governing the collection of DNA from arrestees. Sometimes a court order is required to retrieve a reference from a person of interest. Reference samples are always collected from victims unless they choose not to cooperate with the investigation; in that case, a court order might be required.

Reference samples are often collected by swabbing the inside of the cheek.
In addition to unknown and reference samples, elimination samples are often collected from consensual sex partners and others, such as first responders, crime scene personnel and analysts working the case so they can be excluded from the investigation.
It is important that biological evidence be properly collected and preserved as it can easily degrade when exposed to heat or humidity. Storing evidence in cool environments is preferred; however, research has shown that room temperature conditions are suitable for storing dried stains as long as the humidity is controlled. Liquid samples should be transported in refrigerated or insulated containers.
Who Conducts DNA Analysis
DNA analysts working in laboratories that participate in the FBI’s National DNA Index System (NDIS) and/or are accredited by a recognized organization must meet specific educational and training requirements. At a minimum, a bachelor’s degree in biology, chemistry, or a forensic science-related area is required. In addition, the analyst should have successfully completed nine hours of coursework at the undergraduate or graduate level covering the following subject areas: biochemistry, genetics, molecular biology, as well as coursework or training in statistics and/or population genetics, as it applies to forensic DNA analysis.
To ensure analysts’ skills are kept up to date, analysts who are actively employed at a crime laboratory are also required to meet continuing education requirements. These requirements are stipulated by the FBI’s Quality Assurance Standards (QAS) .
The specialists who conduct DNA analysis in the laboratory are referred to by several different titles, including: Crime Laboratory Analyst, Forensic Examiner, Forensic Scientist and Forensic Laboratory Analyst.
How and Where DNA Testing is Performed
DNA testing must be conducted in a laboratory with dedicated facilities and equipment that meet the FBI’s stringent QAS requirements . Most publicly funded DNA crime laboratories in the United States are part of state, regional or municipal law enforcement agencies and accept submissions from multiple agencies.
Prior to performing DNA analysis at the laboratory, initial testing is often conducted at the crime scene to determine the type of biological material in question. Screening for the presence of biological materials may also be conducted in the laboratory to determine if a specific biological fluid may be present. Most biological screening tests are presumptive in nature and do not specifically identify a bodily fluid.
To determine who deposited biological material at a crime scene, unknown samples are collected and then compared to known samples taken directly from a suspect or victim.
Most DNA samples submitted to a laboratory undergo the following process:
- Extraction is the process of releasing the DNA from the cell.
- Quantitation is the process of determining how much DNA you have.
- Amplification is the process of producing multiple copies of the DNA in order to characterize it.
- Separation is the process of separating amplified DNA product to permit subsequent identification.
- Analysis & Interpretation is the process of quantitatively and qualitatively comparing DNA evidence samples to known DNA profiles.
- Quality Assurance is the process of reviewing analyst reports for technical accuracy.

During extraction, a centrifuge is used to concentrate the sample to the base of the tube.
How the Results are Interpreted
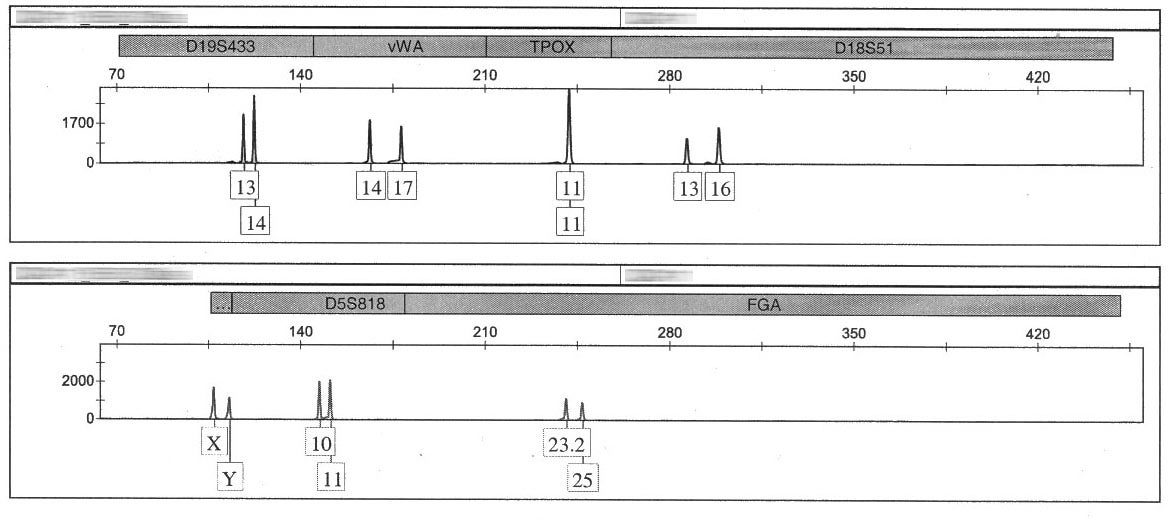
The DNA analysis process provides the analyst with a chart called an electropherogram, which displays the genetic material present at each loci tested (each of the gray bars on the graph below, except for the last one, correspond to a locus; the final gray area is used to indicate the gender of the individual). In a complete profile, each person will exhibit either one or two peaks (alleles) at each locus. The following electropherogram is an example of a profile from a single individual (i.e., a “single-source” profile):

Click image to view larger.
Loci that display only one allele indicate that the individual inherited the same marker from both parents at this locus. Where two alleles are displayed, the individual inherited different markers.

This image shows that the first four loci from the unknown evidence sample collected at the scene match the sample collected from the suspect. (This process would be repeated for all 13 loci.)
Note : The height of each peak must exceed a predetermined quantity threshold to be used in the analysis.
Is the suspect included?
In practice, evidence often contains a mixture of DNA from more than one person. These mixtures can be very challenging to analyze and interpret. In the following example, each marker from the suspect sample is included in the mixture profile collected from the evidence.

Partial Profiles:
If any locus is missing an allele, this is considered a partial profile. Partial profiles can happen for a variety of reasons, such as when a sample is degraded. If a sample has peaks at every locus, but any of them fall below a predetermined threshold, this would also be considered a partial profile.

The partial DNA profile displayed above is missing peaks at two loci. Click image to view larger.
Comparing Profiles Against a Central Database
To enable profiles to be searched against a large, national database, the FBI created the National DNA Index System (NDIS) in 1998. This national database is part of the Combined DNA Index System (CODIS) that enables law enforcement agencies throughout the nation to share and compare DNA profiles to help investigate cases. As of 2012, there are more than 10 million DNA profiles in the system and CODIS has produced leads that have assisted in almost 170,000 investigations .
Once a laboratory enters a case into CODIS, a weekly search is conducted of the DNA profiles in NDIS, and resulting matches are automatically returned to the laboratory that originally submitted the DNA profile. CODIS has three levels of operation:
- Local DNA Index System(LDIS)
- State DNA Index System (SDIS)
- National DNA Index System (NDIS)

The NDIS databases contain DNA profiles from:
- Convicted Offenders - DNA profiles of individuals convicted of crimes
- Arrestees - profiles of arrested persons (if state law permits the collection of arrestee samples)
- Forensic unknowns - DNA profiles of unknown individuals developed from crime scene evidence, such as semen stains or blood
- Missing Persons - contains DNA reference profiles from missing persons
- Biological Relatives of Missing Persons - contains DNA profiles voluntarily contributed from relatives of missing persons
- Unidentified Humans (Remains) - contains DNA profiles developed from unidentified human remains
Each database has its own rules regarding the number of STR markers that must be present for the profile to be uploaded. As of January 1, 2017, the National DNA Index System (NDIS) requires that 20 autosomal STR markers be tested, and the profile must contain information for at least 10 loci. The requirements are less stringent for state and local databases. States require the profile to have information for seven or more loci, and the local database requires at least four loci to be present to be uploaded.
Back to top of page ▲

Find Out More
- Introduction
- Applications
- How It’s Done ◀
- Common Terms
- Resources & References
- International edition
- Australia edition
- Europe edition

Killer breakthrough – the day DNA evidence first nailed a murderer
It’s 30 years since DNA fingerprinting was first used in a police investigation. The technique has since put millions of criminals behind bars – and it all began when one scientist stumbled on the idea in a failed experiment
T hirty years ago this summer, at 4.30 one Thursday afternoon, a 15-year-old schoolgirl called Dawn Ashworth set off from a friend’s house in the village of Narborough, Leicestershire, and began to walk home. Dawn lived in the nearby village of Enderby, a few minutes’ walk away, and chose to take a short-cut along a footpath known locally as Ten Pound Lane. And then she vanished. It was not until two days later that Dawn’s body was found in the corner of a nearby field, covered in twigs, branches and torn-up nettles. The pathologist established that she had put up a considerable struggle before being raped and strangled.
The hunt for Dawn’s killer was unlike any previous murder investigation, however: it was conducted with the help of a new science. The technique known as DNA fingerprinting was employed in a criminal investigation for the first time. Not only did this revolutionary technique lead, indirectly, to the killer being caught; it also prevented a grave miscarriage of justice. And it was employed in a manner that would, today, be likely to face resistance from some members of the public.
As soon as Dawn’s body was found, the police realised that they were looking for a serial killer: two-and-a-half years earlier, another 15-year-old, Lynda Mann, had been murdered a few hundred yards from the scene of Dawn’s murder. Lynda’s clothes had been removed in the same manner as Dawn’s, and she too had been raped before being strangled with her own scarf. Detectives also believed that the killer was a local man, someone who knew the area, and possibly knew Lynda.
Narborough and the surrounding villages were paralysed with fear. In an editorial headlined “Killer in our midst”, the Leicester Mercury warned: “If we don’t catch him it could be your daughter next.” The vicar of Enderby, Canon Alan Green, urged the murderer to give himself up, “because at some time in the future you will have to face your creator and account for the terrible thing you have done”.
While the police had been unable to find Lynda’s killer, they quickly made an arrest after the discovery of Dawn’s body.

Richard Buckland, a 17-year-old Narborough boy with learning difficulties, and who knew Dawn, appeared to have knowledge of some details of the crime that had not been made public. Under questioning he would repeatedly admit the crime, and then withdraw the admission. On 10 August he was charged with Dawn’s murder, and appeared in court the following day.
Buckland refused to confess to the murder of Lynda, however. He was adamant that he was not guilty of that crime. The police, in their certainty that both girls’ lives had been taken by the same person, were convinced he was lying.
Meanwhile, five miles north-west of Narborough, at the University of Leicester, geneticist Alec Jeffreys had made a remarkable – and quite accidental – discovery during a failed experiment to study the way in which inherited illnesses pass through families. He had extracted DNA from cells and attached it to photographic film, which was then left in a photographic developing tank. Once extracted, the film showed a sequence of bars: Jeffreys quickly realised that every individual whose cells had been used in the experiment could be identified with great precision . Furthermore, the technique could be used to determine kinship.
After publishing an academic paper on his discovery, Jeffreys had been asked to assist with a number of cases in which children were being denied British citizenship because immigration officials were disputing that they were the offspring of British parents. But DNA fingerprinting had yet to be used in a criminal investigation. When Jeffreys gave his first talk about his discovery and suggested that it could be used to apprehend criminals, some in the audience had laughed out loud.
But after Buckland’s court appearance, Jeffreys received an unexpected call from the police, asking him whether his new science could prove that the youth had murdered Lynda as well as Dawn.
Jeffreys agreed to carry out tests on Buckland’s blood and on semen taken from the dead girls’ bodies, and worked through the night to finish the work. When he took the film from the developing tank, he could see immediately that the girls had been raped by the same man – and also that Buckland’s DNA was completely different. He had to tell the police that although they were correct in their belief that one man had raped and murdered both girls, not only had Buckland not killed Lynda, he had not killed Dawn either.

The police were astonished, and initially reluctant to believe what they were hearing. Jeffreys’ test was repeated, and then run a third time. “One minute we got the guy,” the senior investigating officer is said to have muttered, “and the next we’ve got Jack shit.”
Buckland, clearly innocent, was set free – after more than three months in custody – and the police were back at square one in their hunt for a highly-dangerous double killer.
The following month, the detectives decided that the technology that had exonerated Buckland should be used to catch the killer. They would screen the entire neighbourhood: they would set up an operation to gather the DNA of every man in the area. Letters were sent to every male born between 1953 and 1970, who had lived or worked in the Narborough area in recent years, asking them to agree to give a blood sample. Two testing centres were established, in a local school and a council office, and there were two testing sessions, morning and evening, three days a week. Each man was expected to bring proof of identity.
It was a voluntary scheme, and a few men declined, some saying they did not like needles, one or two saying they did not like police officers. But most of these men soon changed their minds. The horror at the crimes – and the fear that the killer could strike again – resulted in those with reservations coming under considerable social pressure. By the end of the month, around 1,000 men had volunteered to give samples, and the forensic science laboratories that were conducting the tests were struggling to keep up.
The process quickly drew national and international attention. “Police investigating the murders of two teenage girls near this small Midlands village are applying a new scientific technique,” reported the Los Angeles Times. “Some predict [it] could be the most significant breakthrough in resolving serious crime since fingerprinting was invented.”

There were complaints from some, with the National Council for Civil Liberties – as the UK rights group Liberty was formerly known – highlighting the risk of human error and suggesting that parliament needed to consider the implications of mass screening programmes. But as the LA Times put it: “A strong sense of community outrage among close-knit villagers and an effective police public relations campaign effectively overcame apprehensions among some residents that the tests were an invasion of their personal rights.”
After eight months, 5,511 men had given blood samples and only one had refused. But there was no match with the semen samples. Police began to expand the hunt.
Among those who were recorded as having given a sample was Colin Pitchfork, a 27-year-old baker and father of two young children. Three years earlier, he had been questioned about his movements on the evening that Lynda had been murdered. He had said, quite correctly, that he had been looking after his young son.
In August 1987, more than a year after the killing of Dawn, one of Pitchfork’s workmates, a man called Kelly, was having a pint with a few friends in a pub in Leicester. The conversation turned to Pitchfork, and Kelly confessed that he had impersonated him, in order to take the blood test on his behalf. Kelly explained that Pitchfork had asked for this favour because he had already taken the test, for a friend who had a conviction for indecent exposure when he was younger. Pitchfork had doctored his passport, inserting Kelly’s photograph, and then driven him to the test centre at the school, waiting outside while the blood sample was taken.
Six weeks later, one of the people in the pub relayed this conversation to a local policeman. Kelly was promptly arrested, and by the end of the day Pitchfork was also in custody. After reading him his rights, a detective asked: “Why Dawn Ashworth?”
Pitchfork is said to have shrugged and replied: “Opportunity. She was there and I was there.”

He then gave a detailed confession to both murders and two other sexual assaults. When he raped and killed Lynda Mann, his car had been parked nearby, and his baby son had been asleep in the back of it. DNA testing confirmed him as the double killer.
The following January Pitchfork appeared at Leicester crown court, where he pleaded guilty to two counts of murder, two of rape, two of indecent assault and one count of conspiring to pervert the course of justice.
The court was provided with a psychiatric report that recorded a “personality disorder of psychopathic type accompanied by serious psychosexual pathology” and warned that Pitchfork “will obviously continue to be an extremely dangerous individual while the psychopathology continues”. He was sentenced to life imprisonment, and in accordance with the practice at that time, the home secretary, Douglas Hurd, set his minimum term: 30 years .
Although Jeffreys’ work almost certainly saved Buckland from suffering a serious miscarriage of justice, Pitchfork’s guilty pleas meant that DNA evidence was not relied upon by the prosecution, and this new science was not tested by the court. But the investigative potential of the technique was rapidly recognised and embraced by police forces around the globe. The Home Office took immediate steps to ensure sufficient numbers of technicians and forensic scientists were trained to allow DNA profiling to be incorporated routinely into police casework.
Over the last 30 years, according to some estimates, more than 50 million people have had their DNA tested during criminal investigations. It has secured the convictions of perhaps millions of criminals and, from time to time, excluded innocent suspects like Buckland, and overturned miscarriages of justice.
There have also been occasional mass DNA dragnets in Australia , Canada , France , Germany, the Netherlands, the US and elsewhere, but in recent years, police forces have been less likely to use the mass screening programmes of the type that led, indirectly, to the apprehension of Pitchfork.

The Leicestershire detectives conceded at the time of their DNA sweep that while they were able to persuade thousands living in a small – and shocked – rural community to co-operate, they would not have encountered the same response if the murders had happened in a city.
Jeremy Gans , a professor at Melbourne Law School who has studied DNA dragnets, believes the main reason such sweeps are now uncommon may be because many countries have created large, standing DNA databases. “The police, like almost everyone else, think that if you’re looking for someone who has committed a serious crime, you are looking for someone who has had prior contact with the police.”
The first national DNA database was established in the UK in 1995 . It currently holds 5.8 million DNA profiles based on samples taken from 5.1 million people – equivalent to almost 8% of the population.
Gans says that other reasons why DNA sweeps may no longer be deemed useful include the ease with which police can obtain DNA from suspects – such as by following them until they discard a soft drink can – and the development of the technique to the point where a match to a crime scene sample can be made through samples taken from relatives of the criminal.
It is also possible that police forces in the UK became less interested in gathering large numbers of samples following the introduction of new legislation in 2012 that provided for the eventual destruction of most DNA profiles based on samples taken from people who were not convicted of a crime. The law was introduced to address concerns that the retention of the DNA records of everyone who was arrested - regardless of whether they were eventually charged and prosecuted - risked undermining public support for the national DNA database.
Alec – by now Professor – Jeffreys had supported this reform. By this time, he had became known as the father of genetic fingerprinting. In 1994 he was knighted for services to science and technology and four years ago he retired from the university.
Friends of Lynda and Dawn, meanwhile, have set up a web page in memory of the girls . People from the area add their recollections, and use the site to lobby against the release of Pitchfork.
In prison, Pitchfork is said to have been well behaved, has been educated to degree level, and has become a specialist in the transcription of printed music into Braille. In April this year he appeared before the parole board, which recommended that he be moved to an open prison, but not released.
Not surprisingly, Lynda’s mother, Kath Eastwood, was unimpressed. “He may have a degree,” she told the Mercury, “but he is also a double child murderer and rapist. You can say he is a well-behaved prisoner, but don’t ever forget that he is a well-behaved double child killer.”
Strands of evidence: famous DNA cases

In 1990 Alec Jeffreys established that DNA taken from a femur recovered from a Brazilian graveyard was almost certainly that of Josef Mengele , a former SS officer and physician who subjected Auschwitz inmates to grotesque medical experiments. The match was made with samples taken from Mengele’s relatives, who declined to accept his remains.
A year later scientists were able to prove that skeletal remains found in a burial pit in Yekaterinburg, 850 miles east of Moscow, were those of Russia’s last tsar, Nicholas II , and his wife and children. They had all been killed in 1918, during the Russian civil war. Matches were made with samples taken from several people related to the family, including the Duke of Edinburgh.
In the US, at least 17 death row inmates have been freed after being exonerated by new DNA evidence. One of them, Henry McCollum , had served 31 years behind bars.
Familial DNA matches are increasingly bringing criminals to justice long after the crimes were committed. Last month Christopher Hampton , 64, was jailed for life at Bristol crown court after admitting that he had raped a 17-year-old girl and stabbed her to death in 1984. Samples taken from the crime scene were found to match DNA taken from Hampton’s daughter after she was arrested during a domestic dispute.
- DNA database
Comments (…)
Most viewed.
- Reference Manager
- Simple TEXT file
People also looked at
Mini review article, past, present, and future of dna typing for analyzing human and non-human forensic samples.

- 1 Department of Biological Sciences, Florida International University, Miami, FL, United States
- 2 International Forensic Research Institute, Florida International University, Miami, FL, United States
Forensic DNA analysis has vastly evolved since the first forensic samples were evaluated by restriction fragment length polymorphism (RFLP). Methodologies advanced from gel electrophoresis techniques to capillary electrophoresis and now to next generation sequencing (NGS). Capillary electrophoresis was and still is the standard method used in forensic analysis. However, dependent upon the information needed, there are several different techniques that can be used to type a DNA fragment. Short tandem repeat (STR) fragment analysis, Sanger sequencing, SNapShot, and capillary electrophoresis-single strand conformation polymorphism (CE-SSCP) are a few of the techniques that have been used for the genetic analysis of DNA samples. NGS is the newest and most revolutionary technology and has the potential to be the next standard for genetic analysis. This review briefly encompasses many of the techniques and applications that have been utilized for the analysis of human and nonhuman DNA samples.
Introduction
Forensic genetics applies genetic tools and scientific methodology to solve criminal and civil litigations ( Editorial, 2007 ). Locard’s Exchange Principle states that every contact leaves a trace, making any evidence a key component in forensic analysis. Biological evidence can comprise of cellular material or cell-free DNA from crime scenes, and as technologies improved, genetic methodologies were expanded to include human and non-human forensic analyses. Although these methodologies can be used for any genome, the prevalence of databases and standard guidelines has allowed human DNA typing to become the gold standard. This review will discuss the historical progression of DNA analysis techniques, strengths and limitations, and their possible forensic applications applied to human and non-human genetics.
Methodologies to Detect Genetic Differences in Humans Is the “Gold Standard”
“dna fingerprinting”: the beginning of human forensic dna typing.
“DNA fingerprinting” was serendipitously discovered in 1984 ( Jeffreys, 2013 ). What they found propelled DNA “fingerprinting,” or DNA typing, to the forefront in legal cases to become the “gold standard” for forensic genetics in a court of law. Jeffreys first used restriction enzymes to fragment DNA, a method in which restriction endonucleases (RE) enzymes fragment the genomic DNA, producing restriction fragment length polymorphisms (RFLP) patterns. Since each RE recognizes specific DNA sequences to enzymatically cut the DNA, then inherent differences between gene sequences, due to evolutionary changes, will produce different fragment lengths. If the enzyme site is present in one individual but has changed in a different individual, the fragment lengths, once separated and visualized, will differ. While this technique was useful for some studies, Jeffreys did not find it useful for his particular genetic studies. Subsequently when working with the myoglobin gene in seals, he discovered that a short section of that gene – a minisatellite – was conserved and when isolated and cloned could be used to detect inherited genetic lineages as well as individualize a subject. Fragment length separation by electrophoresis, followed by transfer to Southern blot membranes, hybridized with a specific or non-specific complementary isotopic DNA probe, allowed for DNA fragments visualization ( Jeffreys et al., 1985b ). Upon careful analysis, Jeffreys determined that the fragments represented different combinations of DNA repetitive elements, unique to each individual, and could be used to better identify individuals or kinship lineages ( Jeffreys et al., 1985b ). Jeffreys’ technology was used in several subsequent paternity, immigration, and forensic genetics cases ( Gill et al., 1985 ; Jeffreys et al., 1985a ; Evans, 2007 ). This was just the beginning of a whole new era in DNA typing.
Restriction Fragment Length Polymorphism (RFLP) Analysis: The Past
After Jeffreys’ discoveries, many DNA analyses methods involving electrophoretic fragment separation were discovered. Many were based on RFLP principles ( Botstein et al., 1980 ), e.g., amplified fragment length polymorphism (AFLP) ( Vos et al., 1995 ), and terminal restriction fragment length polymorphism (TRFLP) ( Liu et al., 1997 ). Others like length heterogeneity- polymerase chain reaction (LH-PCR) ( Suzuki et al., 1998 ) were based on intrinsic insertions and deletions of bases within specific genetic markers. Sanger sequencing ( Sanger and Coulson, 1975 ), and single-strand conformational polymorphism (SSCP) analysis ( Orita et al., 1989 ), while separated by electrophoresis, are theoretically based on single base sequence changes rather than insertions, deletions or RE site differences. While Jeffrey’s DNA fingerprinting method provided a very high power of discrimination, the main limitations were it was very time-consuming and required at least 10–25 ng of DNA to be successful ( Wyman and White, 1980 ). With these limitations, RFLP was not always feasible for forensic cases.
Short Tandem Repeat (STR) Analysis: The Present
The polymerase chain reaction (PCR) was discovered by Kary Mullis in 1985 and helped transform all DNA analyses ( Mullis et al., 1986 ). The current standard for human DNA typing is short tandem repeat (STR) analysis ( McCord et al., 2019 ). This method amplifies highly polymorphic, repetitive DNA regions by PCR and separates them by amplicon length using capillary electrophoresis. These inheritable markers are a series of 2–7 bases tandemly repeated at a specific locus, often in non-coding genetic regions. Forensic STRs are commonly tetranucleotide repeats ( Goodwin et al., 2011 ), chosen because of their technical robustness and high variation among individuals ( Kim et al., 2015 ). The combined DNA index system (CODIS) uses 20 core STR loci, expanded in 2017, and several commercial kits are available that contain these STRs ( Oostdik et al., 2014 ; Ludeman et al., 2018 ). After amplification, different fluorochromes on each primer set allow for visualization of STRs after deconvolution, creating a STR profile consisting of a combination of genotypes ( Gill et al., 2015 ). This method has become the gold standard for human forensics. Its greatest strength is the standardization of loci used by all laboratories and an extremely large searchable database of genetic profiles. However, some limitations and challenges are faced when dealing with highly degraded or low template DNA samples. To overcome these technical challenges, standardized mini-STR kits have been developed which use shorter versions of the core STRs and can be used in the same manner for forensic cases ( Butler et al., 2007 ; Constantinescu et al., 2012 ). Keep in mind, DNA typing of humans – a single species – is the gold standard because of (a) the concerted scientific effort to standardize loci to analyze, (b) the development of commercial kits that can produce the same results regardless of instrumentation or laboratory performing the work, (c) a compatible and very large database that provides allelic frequencies for all sub-populations of humans, (d) standardized statistical methods used to report the results and (e) many court cases that have accepted human DNA typing evidence in a court of law – setting the precedent for future cases to use DNA typing results.
Methodologies to Detect Genetic Differences in Non-Humans: Past and Present
Amplified fragment length polymorphism (aflp) analysis.
It was not long before scientists realized that non-human DNA could provide informative genetic evidence in forensic cases. Applications include bioterrorism, wildlife crimes, human identification through skin microorganisms, and so much more ( Arenas et al., 2017 ). Since large quantities of biological materials are frequently not found at crime scenes, successful RFLP analyses were unlikely. Combining restriction enzymes and PCR technology, a process known as AFLP analysis ( Vos et al., 1995 ), became a method for DNA fingerprinting using minute amounts of unknown sourced DNA. REs digest genomic DNA, then ligation of a constructed adapter sequence to the ends of all fragments allows the annealing of primers designed to recognize the adaptor sequences. Subsequent amplification generates many amplicons ranging in length when separated and visualized in an electropherogram or on a gel ( Vos et al., 1995 ; Butler, 2012 ). AFLP markers for plant forensic DNA typing have been used because it provides high discrimination, requires only small amounts of DNA and the method is reproducible, all forensically important characteristics ( Datwyler and Weiblen, 2006 ). For example, since most cannabis is clonally propagated, subsequent generations will have identical genetic profiles as seen with AFLP ( Miller Coyle et al., 2003 ), providing useful intelligence links back to the source population. But there are significant variation between cultivars and within populations, so not having a standard database representing the species’ diversity for statistical comparisons greatly limits the method’s applicability. Another forensic example of its use is differentiating between marijuana and hemp, two morphologically and genetically similar plants, one an illicit drug while the other is not. In this study, three populations of hemp and one population of marijuana were analyzed with AFLP producing 18 bands that were specific to hemp samples. Additionally, 51.9% of molecular variance occurred within populations indicating these polymorphisms were useful for forensic individualization ( Datwyler and Weiblen, 2006 ).
Terminal Restriction Fragment Length Polymorphism (TRFLP) Analysis
As a result of the anthrax letter attacks of 2001, microbial forensics came to the forefront ( Schmedes et al., 2016 ), a discipline that combines multiple scientific specialties – microbiology, genetics, forensic science, and analytical chemistry. One method used to compare microbial communities is TRFLP ( Liu et al., 1997 ; Osborn et al., 2000 ; Butler, 2012 ). With this method, the DNA is amplified using “universal,” highly conserved primer sequences shared across all organisms of interest, i.e., the 16S rRNA genes in bacteria and Archaea, and then uses REs to fragment the PCR products ( Table 1 ). Separated by capillary electrophoresis, only the fluorescently tagged terminal restricted fragments are visualized ( Mrkonjic Fuka et al., 2007 ), reducing the profile complexity and providing high discrimination. TRFLP has been used to characterize complex microbial communities for forensic applications by linking the similarity of the amplicon patterns generated from the intrinsic soil communities to the evidence from a crime scene ( Meyers and Foran, 2008 ; Habtom et al., 2017 ). This method does provide a distinct pattern reflective of the microbial community, useful for forensic genetics but the method does not provide any sequence information. Another limitation is no standardization of which primer pairs or REs are used, making direct comparisons between studies difficult. This lack of standardization also hinders the development of a database for species identification. Additionally, the method is time-consuming due to the additional step of restriction digestion and the possibility of incomplete enzymatic digestion can complicate the interpretation of results ( Osborn et al., 2000 ; Moreno et al., 2006 ).
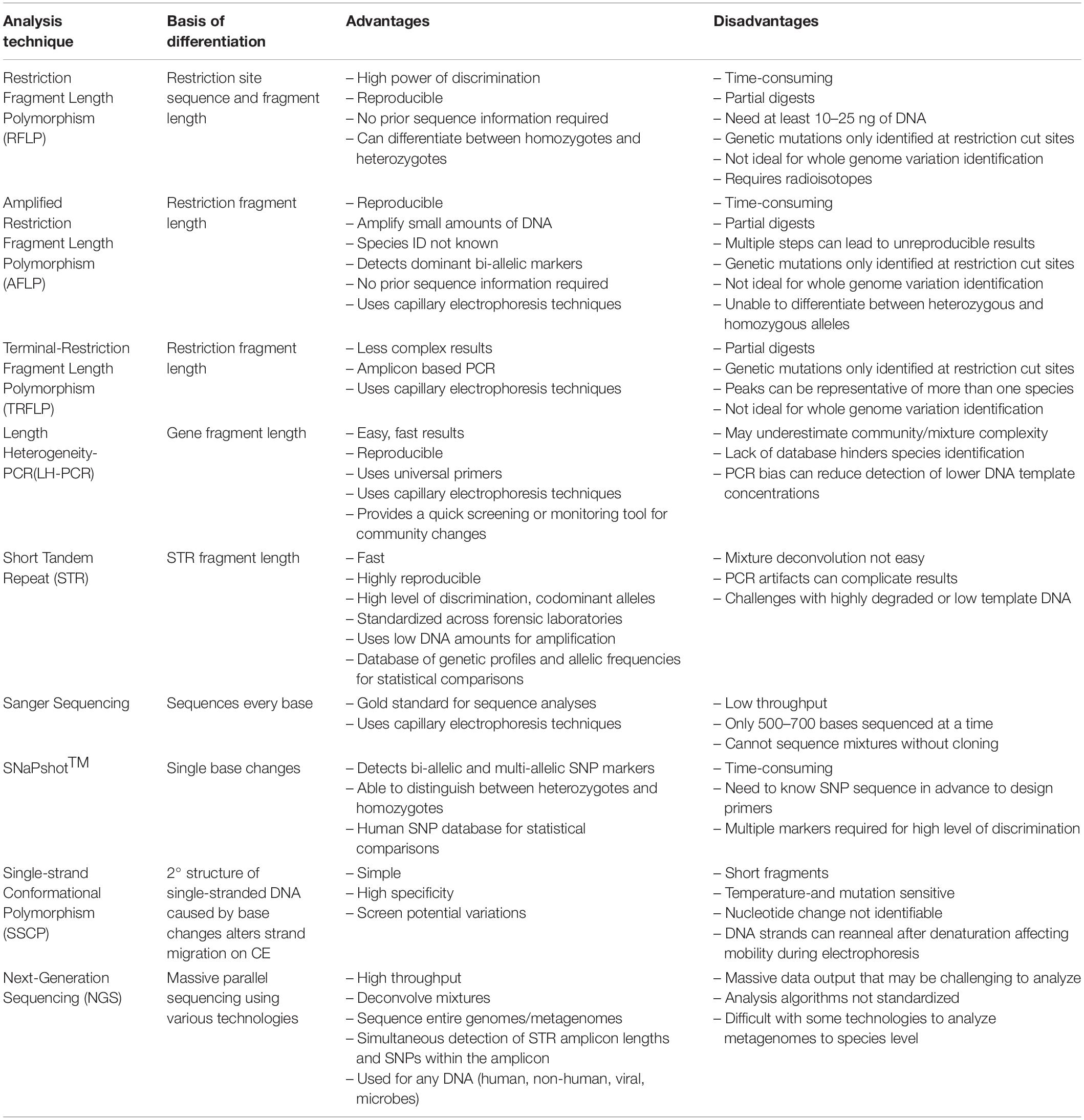
Table 1. The basis of differentiation, advantages, and disadvantages of past and current technologies.
Length Heterogeneity-Polymerase Chain Reaction (LH-PCR)
Another methodology has been used to characterize microbial communities is length heterogeneity- polymerase chain reaction (LH-PCR) ( Suzuki et al., 1998 ). Universal primers complementary to highly conserved domains within genomes are used to amplify hypervariable sequences within specific sequence domains. The 16S/18S rRNA genes, the chloroplast genes or Internal Transcribed Spacer (ITS) regions are commonly used. This technique is based on the natural sequence length variation due to insertions and deletions of bases that occur within a domain ( Moreno et al., 2006 ). It has been used to characterize microbial communities for forensic soil applications where a correlation between geographic location and microbial profiles has proven to be more discriminating than elemental soil analysis ( Moreno et al., 2006 , 2011 ; Damaso et al., 2018 ). With LH-PCR, metagenomic DNA extracted from the soil is amplified using fluorescently labeled universal primers with amplicon peaks within the electropherogram representing the minimum diversity within the community. However, specific sequence information is not known as many peaks of the same size could represent more than one species, thereby masking the community’s actual taxonomic diversity. A recent study showed the intrinsic diversity of a microbial mat, masked by LH-PCR, could be further resolved by the inherent sequence differences using capillary electrophoresis-single strand conformational polymorphism (CE-SSCP) analysis ( Damaso et al., 2014 ) and confirmed by sequencing. The advantage of LH-PCR is it is a fast and reproducible method that can correlate geographical areas to microbial patterns with bioinformatics ( Damaso et al., 2018 ); but a soil database would need to be developed to be useful beyond specific geographical areas.
Methodologies to Detect Intersequence Variation: The Past and Present
Sanger sequencing and single nucleotide polymorphism (snp) variation.
The basis of genomic differentiation is the intrinsic order of base pairs within a region that can be evaluated by sequencing. Sanger sequencing has been the gold standard since the 1970s ( Sanger and Coulson, 1975 ). Sanger sequencing was termed the gold standard because of the ability for single base pair resolution allowing for full sequence information to be determined. Robust and extensive databases are also readily available for comparison, i.e., GenBank, to identify an organism. However, it does have some limitations such as the short length (<500–700 bp) and it cannot sequence mixtures of organisms, for example, without cloning, so it would not be useful for sequencing complex microbial communities without intense time, effort and cost.
Other approaches use the ability to identify intrinsic single base sequence variation using single nucleotide polymorphisms (SNPs) within four forensically relevant SNP classes: identity-testing, ancestry informative, phenotype informative, and lineage informative. SNPs are particularly useful when typing degraded DNA or increasing the amount of genetic information retrieved from a sample ( Budowle and van Daal, 2008 ; Goodwin et al., 2011 ). SNaPshot TM is a commercially available SNP kit that can identify known SNPs using single base extension (SBE) technology ( Daniel et al., 2015 ; Fondevila et al., 2017 ). Wildlife forensics has used SNaPshot TM to identify endangered or trafficked species that are illegally poached to support criminal prosecutions. Elephant species identification from ivory and ivory products ( Kitpipit et al., 2017 ) or differentiating wolf species from dog subspecies ( Jiang et al., 2020 ) are both examples of SNaPshot TM assays developed for wildlife forensics. By using species-specific SNPs, the samples could be identified. But yet again, the limitation becomes the need for species-specific reference databases and the monumental task of developing a robust database for each species. Human SNPs databases with allele frequencies, as seen in dbSNP, however, are available making their forensic application more feasible in some cases.
Next-Generation Sequencing: The Present
Massively parallel sequencing (MPS) or next-generation sequencing (NGS) allows for mixtures of genomes of any species to be sequenced in one analysis ( Ansorge, 2009 ). This technology can sequence thousands of genomic regions simultaneously, allowing for whole-genome, metagenomic sequencing or targeted amplicon sequencing ( Gettings et al., 2016 ). Various NGS technologies are available each using slightly different technologies to sequence DNA ( Heather and Chain, 2016 ). Verogen has developed kits explicitly for human forensic genomics using Illumina’s MiSeq FGx system ( Guo et al., 2017 ; Moreno et al., 2018 ). The FBI recently approved DNA profiles generated by Verogen forensic technology to be uploaded into the National DNA Index System (NDIS) ( SWGDAM, 2019 ), making it the first NGS technology approved for NDIS.
Short tandem repeat mixture deconvolution, degraded, low template samples, and even microbial community samples are just a few of the potential NGS applications for forensic genomics and metagenomics ( Borsting and Morling, 2015 ). In human STR analyses, the greatest challenge is mixture deconvolution. NGS technology presents an increased power of discrimination of STR alleles using the intrinsic SNPs genetic microhaplotypes – a combination of 2–4 closely linked SNPs within an allele ( Kidd et al., 2014 ; Pang et al., 2020 ). However, the acceptance of analyses programs to deconvolve mixtures has not been standardized to the same level as it has for STRs.
Microbes are the first responders to changes in any environment because they are rapidly affected by the availability of nutrients and their intrinsic habitats. This makes them excellent indicators for studies investigating post-mortem interval (PMI) or as an indicator of soil geographical provenance ( Giampaoli et al., 2014 ; Finley et al., 2015 ). In decaying organisms, shifts in epinecrotic communities or the thanatomicrobiome are becoming increasingly critical components in investigating PMI ( Javan et al., 2016 ). Sequencing of the thanatomicrobiome revealed the Clostridium spp. varied during different stages human decomposition, the “Postmortem Clostridium Effect” (PCE), providing a time signature of the thanatomicrobiome, which could only have been uncovered through NGS ( Javan et al., 2017 ). However, the lack of consensus in analyses techniques must be addressed before NGS methodologies can be introduced into the justice system ( Table 1 ).
Future Directions and Concluding Remarks
Forensic DNA typing has progressed quickly within a short timeframe ( Figure 1 ), which can be attributed to the many advancements in molecular biology technologies. As these techniques advance, forensic scientists will analyze more atypical forms of evidence to answer questions deemed unresolvable with traditional DNA analyses. For example, epigenetics and DNA methylation markers have been proposed to estimate age, determine the tissue type, and even differentiate between monozygotic twins ( Vidaki and Kayser, 2018 ). However, since epigenetic patterns are also influenced by environmental factors, they can be dynamic, and a number of confounding factors have the potential to affect predictions and must be taken into account when preparing prediction models (i.e., age estimation). Additionally, phenotype informative SNPs across the genome can infer physical characteristics like eye, hair, and skin color, even age, from an unknown source of DNA retrieved from a crime scene. But this technology could pose an “implicit bias” toward minorities, especially in “societies where racism and xenophobia are now on the rise” ( Schneider et al., 2019 ) if not ethically and judicially implemented. With the increased sensitivity of NGS, low biomass samples from environmental DNA (eDNA) – DNA from soil, water, air – can complement and enhance intelligence gathering or provenance in criminal cases. Pollen and dust are two types of eDNA recently explored for their future forensic potential ( Alotaibi et al., 2020 ; Young and Linacre, 2021 ). However, if used in criminal investigations where the eDNA collected has had interaction with other environments, there must be some protocol or quality control established to account for variability that is likely to occur. This makes the prudent validation of this type of DNA analysis, essential. Limitations also arise due to lack of a database for comparison of samples and statistical analyses to evaluate the strength of a match like in the analysis of human STR profiles.
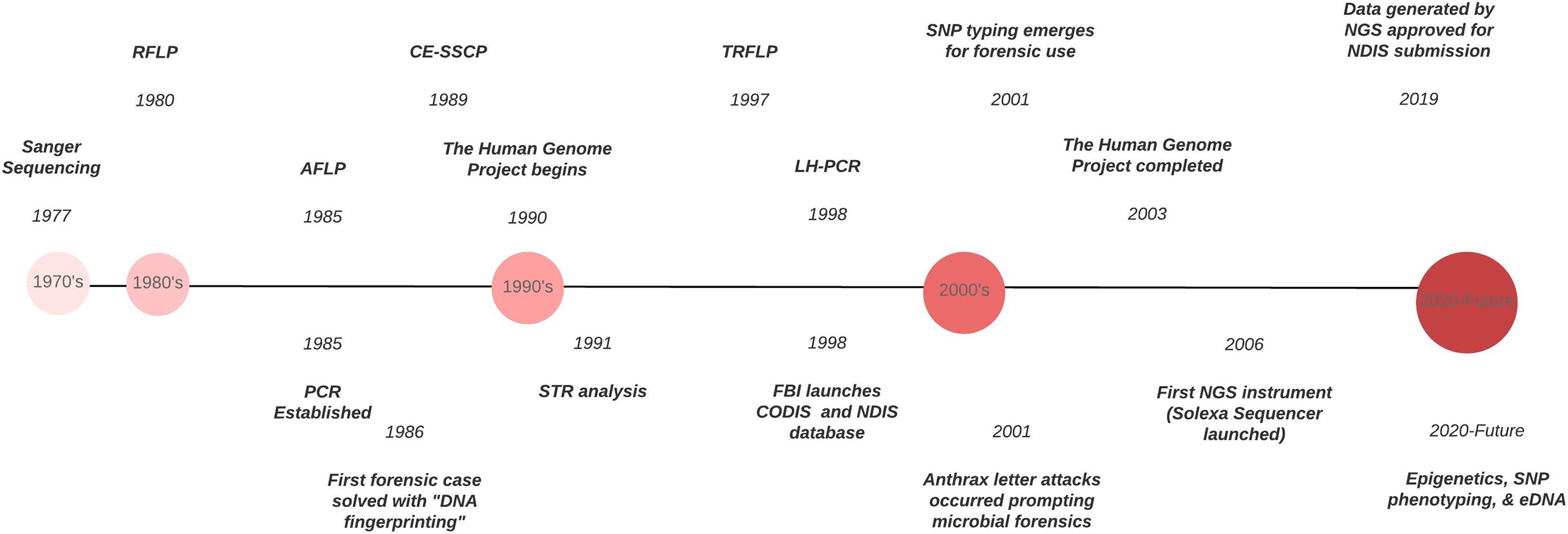
Figure 1. Timeline of the evolution of DNA typing technologies from the 1970’s to the present.
DNA has long been the gold standard in human forensic analysis because of the standardization of DNA markers, databases and statistical analyses. It has laid the foundation for these promising new technologies that will significantly enhance intelligence gathering and species identification – human and non-human – in forensic cases. In order for these methodologies to be useful in criminal investigations, they must adhere to the legal standards such as the Frye or Daubert Standards which determines if an expert testimony or evidence is admissible in court. A method can be deemed acceptable if it follows forensic guidelines set by organizations such as NIST’s Organization Scientific Area Committees (OSAC), Society for Wildlife Forensic Sciences (SWFS), Scientific Working Group on DNA Analysis Methods (SWGDAM), and the International Society for Forensic Genetics (ISFG) ( Linacre et al., 2011 ) just to name a few. These committees provide the guidelines for validation, interpretation, and quality assurance, all necessary components for DNA analysis. The US Fish and Wildlife forensic laboratory has standardized protocols for crimes against federally endangered or threatened species 1 . However, the more common limiting factors in the development of standard guidelines of non-human forensic genetic analyses across different state laboratories are the lack of consensus in methodologies, supporting allelic databases and standardized statistical analyses. Addressing those issues could lay the foundation for non-human analyses to be on par with human analyses.
Author Contributions
DJ designed and wrote the manuscript. DM edited and contributed to the writing of the manuscript. Both authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to acknowledge the invitation by the editors to contribute to this special edition. DJ was supported by the Florida Education Fund’s McKnight Doctoral Fellowship.
- ^ https://www.fws.gov/lab/about.php
Alotaibi, S. S., Sayed, S. M., Alosaimi, M., Alharthi, R., Banjar, A., Abdulqader, N., et al. (2020). Pollen molecular biology: Applications in the forensic palynology and future prospects: A review. Saudi J. Biol. Sci. 27, 1185–1190. doi: 10.1016/j.sjbs.2020.02.019
PubMed Abstract | CrossRef Full Text | Google Scholar
Ansorge, W. J. (2009). Next-generation DNA sequencing techniques. N. Biotechnol. 25, 195–203. doi: 10.1016/j.nbt.2008.12.009
Arenas, M., Pereira, F., Oliveira, M., Pinto, N., Lopes, A. M., Gomes, V., et al. (2017). Forensic genetics and genomics: Much more than just a human affair. PLoS Genet. 13:e1006960. doi: 10.1371/journal.pgen.1006960
Borsting, C., and Morling, N. (2015). Next generation sequencing and its applications in forensic genetics. Forensic Sci. Int. Genet. 18, 78–89. doi: 10.1016/j.fsigen.2015.02.002
Botstein, D., White, R. L., Skolnick, M., and Davis, R. W. (1980). Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 32, 314–331.
Google Scholar
Budowle, B., and van Daal, A. (2008). Forensically relevant SNP classes. Biotechniques 60:610. doi: 10.2144/000112806
Butler, J. M. (2012). “Non-human DNA,” in Advanced Topics in Forensic DNA Typing , ed. J. M. Butler (San Diego: Academic Press), 473–495.
Butler, J. M., Coble, M. D., and Vallone, P. M. (2007). STRs vs. SNPs: thoughts on the future of forensic DNA testing. Forensic Sci. Med. Pathol. 3, 200–205. doi: 10.1007/s12024-007-0018-1
Constantinescu, C. M., Barbarii, L. E., Iancu, C. B., Constantinescu, A., Iancu, D., and Girbea, G. (2012). Challenging DNA samples solved with MiniSTR analysis. Brief overview. Rom. J. Leg. Med. 20, 51–56. doi: 10.4323/rjlm.2012.51
CrossRef Full Text | Google Scholar
Damaso, N., Martin, L., Kushwaha, P., and Mills, D. (2014). F-108 polymer and capillary electrophoresis easily resolves complex environmental DNA mixtures and SNPs. Electrophoresis 35, 3208–3211. doi: 10.1002/elps.201400069
Damaso, N., Mendel, J., Mendoza, M., von Wettberg, E. J., Narasimhan, G., and Mills, D. (2018). Bioinformatics Approach to Assess the Biogeographical Patterns of Soil Communities: The Utility for Soil Provenance. J. Forensic. Sci. 63, 1033–1042. doi: 10.1111/1556-4029.13741
Daniel, R., Santos, C., Phillips, C., Fondevila, M., van Oorschot, R. A., Carracedo, A., et al. (2015). A SNaPshot of next generation sequencing for forensic SNP analysis. Forensic Sci. Int. Genet. 14, 50–60. doi: 10.1016/j.fsigen.2014.08.013
Datwyler, S. L., and Weiblen, G. D. (2006). Genetic variation in hemp and marijuana (Cannabis sativa L.) according to amplified fragment length polymorphisms. J. Forensic Sci. 51, 371–375. doi: 10.1111/j.1556-4029.2006.00061.x
Editorial. (2007). Launching Forensic Science International daughter journal in 2007: Forensic Science International: Genetics. Forensic Sci. Int. Genet. 1, 1–2. doi: 10.1016/j.fsigen.2006.10.001
Evans, C. (2007). The Casebook of Forensic Detection: How Science Solved 100 of the World’s Most Baffling Crimes. New York, NY: Berkley Books.
Finley, S. J., Benbow, M. E., and Javan, G. T. (2015). Potential applications of soil microbial ecology and next-generation sequencing in criminal investigations. Appl. Soil. Ecol. 88, 69–78. doi: 10.1016/j.apsoil.2015.01.001
Fondevila, M., Borsting, C., Phillips, C., de la Puente, M., Consortium, E. N., Carracedo, A., et al. (2017). Forensic SNP genotyping with SNaPshot: Technical considerations for the development and optimization of multiplexed SNP assays. Forensic Sci. Rev. 29, 57–76.
Gettings, K. B., Kiesler, K. M., Faith, S. A., Montano, E., Baker, C. H., Young, B. A., et al. (2016). Sequence variation of 22 autosomal STR loci detected by next generation sequencing. Forensic Sci. Int. Genet. 21, 15–21. doi: 10.1016/j.fsigen.2015.11.005
Giampaoli, S., Berti, A., Di Maggio, R. M., Pilli, E., Valentini, A., Valeriani, F., et al. (2014). The environmental biological signature: NGS profiling for forensic comparison of soils. Forensic Sci. Int. 240, 41–47. doi: 10.1016/j.forsciint.2014.02.028
Gill, P., Haned, H., Bleka, O., Hansson, O., Dorum, G., and Egeland, T. (2015). Genotyping and interpretation of STR-DNA: Low-template, mixtures and database matches-Twenty years of research and development. Forensic Sci. Int. Genet. 18, 100–117. doi: 10.1016/j.fsigen.2015.03.014
Gill, P., Jeffreys, A. J., and Werrett, D. J. (1985). Forensic application of DNA ‘fingerprints’. Nature 318, 577–579. doi: 10.1038/318577a0
Goodwin, W., Linacre, A., and Hadi, S. (2011). “An introduction to forensic genetics.” 2nd ed. West Sussex, UK: Wiley-Blackwell, 53–62.
Guo, F., Yu, J., Zhang, L., and Li, J. (2017). Massively parallel sequencing of forensic STRs and SNPs using the Illumina((R)) ForenSeq DNA Signature Prep Kit on the MiSeq FGx Forensic Genomics System. Forensic Sci. Int. Genet. 31, 135–148. doi: 10.1016/j.fsigen.2017.09.003
Habtom, H., Demaneche, S., Dawson, L., Azulay, C., Matan, O., Robe, P., et al. (2017). Soil characterisation by bacterial community analysis for forensic applications: A quantitative comparison of environmental technologies. Forensic Sci. Int. Genet. 26, 21–29. doi: 10.1016/j.fsigen.2016.10.005
Heather, J. M., and Chain, B. (2016). The sequence of sequencers: The history of sequencing DNA. Genomics 107, 1–8. doi: 10.1016/j.ygeno.2015.11.003
Javan, G. T., Finley, S. J., Abidin, Z., and Mulle, J. G. (2016). The Thanatomicrobiome: A Missing Piece of the Microbial Puzzle of Death. Front. Microbiol. 7:225. doi: 10.3389/fmicb.2016.00225
Javan, G. T., Finley, S. J., Smith, T., Miller, J., and Wilkinson, J. E. (2017). Cadaver Thanatomicrobiome Signatures: The Ubiquitous Nature of Clostridium Species in Human Decomposition. Front. Microbiol. 8:2096. doi: 10.3389/fmicb.2017.02096
Jeffreys, A. J. (2013). The man behind the DNA fingerprints: an interview with Professor Sir Alec Jeffreys. Investig. Genet. 4:21. doi: 10.1186/2041-2223-4-21
Jeffreys, A. J., Brookfield, J. F., and Semeonoff, R. (1985a). Positive identification of an immigration test-case using human DNA fingerprints. Nature 317, 818–819. doi: 10.1038/317818a0
Jeffreys, A. J., Wilson, V., and Thein, S. L. (1985b). Hypervariable ‘minisatellite’ regions in human DNA. Nature 314, 67–73. doi: 10.1038/314067a0
Jiang, H. H., Li, B., Ma, Y., Bai, S. Y., Dahmer, T. D., Linacre, A., et al. (2020). Forensic validation of a panel of 12 SNPs for identification of Mongolian wolf and dog. Sci. Rep. 10:13249. doi: 10.1038/s41598-020-70225-5
Kidd, K. K., Pakstis, A. J., Speed, W. C., Lagace, R., Chang, J., Wootton, S., et al. (2014). Current sequencing technology makes microhaplotypes a powerful new type of genetic marker for forensics. Forensic Sci. Int. Genet. 12, 215–224. doi: 10.1016/j.fsigen.2014.06.014
Kim, Y. T., Heo, H. Y., Oh, S. H., Lee, S. H., Kim, D. H., and Seo, T. S. (2015). Microchip-based forensic short tandem repeat genotyping. Electrophoresis 36, 1728–1737. doi: 10.1002/elps.201400477
Kitpipit, T., Thongjued, K., Penchart, K., Ouithavon, K., and Chotigeat, W. (2017). Mini-SNaPshot multiplex assays authenticate elephant ivory and simultaneously identify the species origin. Forensic Sci. Int. Genet. 27, 106–115. doi: 10.1016/j.fsigen.2016.12.007
Linacre, A., Gusmão, L., Hecht, W., Hellmann, A. P., Mayr, W. R., Parson, W., et al. (2011). ISFG: Recommendations regarding the use of non-human (animal) DNA in forensic genetic investigations. Forensic Sci. Int. Genet. 5, 501–505. doi: 10.1016/j.fsigen.2010.10.017
Liu, W. T., Marsh, T. L., Cheng, H., and Forney, L. J. (1997). Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63, 4516–4522. doi: 10.1128/AEM.63.11.4516-4522.1997
Ludeman, M. J., Zhong, C., Mulero, J. J., Lagace, R. E., Hennessy, L. K., Short, M. L., et al. (2018). Developmental validation of GlobalFiler PCR amplification kit: a 6-dye multiplex assay designed for amplification of casework samples. Int. J. Legal. Med. 132, 1555–1573. doi: 10.1007/s00414-018-1817-5
McCord, B. R., Gauthier, Q., Cho, S., Roig, M. N., Gibson-Daw, G. C., Young, B., et al. (2019). Forensic DNA Analysis. Anal. Chem. 91, 673–688. doi: 10.1021/acs.analchem.8b05318
Meyers, M. S., and Foran, D. R. (2008). Spatial and temporal influences on bacterial profiling of forensic soil samples. J. Forensic Sci. 53, 652–660. doi: 10.1111/j.1556-4029.2008.00728.x
Miller Coyle, H., Palmbach, T., Juliano, N., Ladd, C., and Lee, H. C. (2003). An overview of DNA methods for the identification and individualization of marijuana. Croat Med. J. 44, 315–321.
Moreno, L. I., Galusha, M. B., and Just, R. (2018). A closer look at Verogen’s Forenseq DNA Signature Prep kit autosomal and Y-STR data for streamlined analysis of routine reference samples. Electrophoresis 39, 2685–2693. doi: 10.1002/elps.201800087
Moreno, L. I., Mills, D. K., Entry, J., Sautter, R. T., and Mathee, K. (2006). Microbial metagenome profiling using amplicon length heterogeneity-polymerase chain reaction proves more effective than elemental analysis in discriminating soil specimens. J. Forensic Sci. 51, 1315–1322. doi: 10.1111/j.1556-4029.2006.00264.x
Moreno, L. I., Mills, D., Fetscher, J., John-Williams, K., Meadows-Jantz, L., and McCord, B. (2011). The application of amplicon length heterogeneity PCR (LH-PCR) for monitoring the dynamics of soil microbial communities associated with cadaver decomposition. J. Microbiol. Methods 84, 388–393. doi: 10.1016/j.mimet.2010.11.023
Mrkonjic Fuka, M., Gesche Braker, S. H., and Philippot, L. (2007). “Molecular Tools to Assess the Diversity and Density of Denitrifying Bacteria in Their Habitats,” in Biology of the Nitrogen Cycle , eds H. Bothe, S. J. Ferguson, and W. E. Newton (Amsterdam: Elsevier), 313–330.
Mullis, K., Faloona, F., Scharf, S., Saiki, R., Horn, G., and Erlich, H. (1986). Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb. Symp. Quant. Biol. 1, 263–273. doi: 10.1101/sqb.1986.051.01.032
Oostdik, K., Lenz, K., Nye, J., Schelling, K., Yet, D., Bruski, S., et al. (2014). Developmental validation of the PowerPlex((R)) Fusion System for analysis of casework and reference samples: A 24-locus multiplex for new database standards. Forensic Sci. Int. Genet. 12, 69–76. doi: 10.1016/j.fsigen.2014.04.013
Orita, M., Iwahana, H., Kanazawa, H., Hayashi, K., and Sekiya, T. (1989). Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc. Natl. Acad. Sci. U S A 86, 2766–2770.
Osborn, A. M., Moore, E. R., and Timmis, K. N. (2000). An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ. Microbiol. 2, 39–50. doi: 10.1046/j.1462-2920.2000.00081.x
Pang, J. B., Rao, M., Chen, Q. F., Ji, A. Q., Zhang, C., Kang, K. L., et al. (2020). A 124-plex Microhaplotype Panel Based on Next-generation Sequencing Developed for Forensic Applications. Sci. Rep. 10:1945. doi: 10.1038/s41598-020-58980-x
Sanger, F., and Coulson, A. R. (1975). A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J. Mol. Biol. 94, 441–448. doi: 10.1016/0022-2836(75)90213-2
Schmedes, S. E., Sajantila, A., and Budowle, B. (2016). Expansion of Microbial Forensics. J. Clin. Microbiol. 54, 1964–1974. doi: 10.1128/JCM.00046-16
Schneider, P. M., Prainsack, B., and Kayser, M. (2019). The Use of Forensic DNA Phenotyping in Predicting Appearance and Biogeographic Ancestry. Dtsch Arztebl. Int. 52, 873–880. doi: 10.3238/arztebl.2019.0873
Suzuki, M., Rappe, M. S., and Giovannoni, S. J. (1998). Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity. Appl. Environ. Microbiol. 64, 4522–4529.
SWGDAM (2019). Addendum to SWGDAM Autosomal Interpretation Guidelines for NGS.Swgdam. Available online at: https://www.swgdam.org/publications
Vidaki, A., and Kayser, M. (2018). Recent progress, methods and perspectives in forensic epigenetics. Forensic Sci. Int. Genet. 37, 180–195. doi: 10.1016/j.fsigen.2018.08.008
Vos, P., Hogers, R., Bleeker, M., Reijans, M., van de Lee, T., Hornes, M., et al. (1995). AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23, 4407–4414. doi: 10.1093/nar/23.21.4407
Wyman, A. R., and White, R. (1980). A highly polymorphic locus in human DNA. Proc. Natl. Acad. Sci. U S A 77, 6754–6758. doi: 10.1073/pnas.77.11.6754
Young, J. M., and Linacre, A. (2021). Massively parallel sequencing is unlocking the potential of environmental trace evidence. Forensic Sci. Int. Genet. 50:102393. doi: 10.1016/j.fsigen.2020.102393
Keywords : forensic genetics, DNA typing, metabarcoding, soil, microbes, minisatellites, next-generation sequencing
Citation: Jordan D and Mills D (2021) Past, Present, and Future of DNA Typing for Analyzing Human and Non-Human Forensic Samples. Front. Ecol. Evol. 9:646130. doi: 10.3389/fevo.2021.646130
Received: 25 December 2020; Accepted: 02 March 2021; Published: 22 March 2021.
Reviewed by:
Copyright © 2021 Jordan and Mills. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY) . The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: DeEtta Mills, [email protected]
This article is part of the Research Topic
Life and Death: New Perspectives and Applications in Forensic Science
UK Edition Change
- UK Politics
- News Videos
- Paris 2024 Olympics
- Rugby Union
- Sport Videos
- John Rentoul
- Mary Dejevsky
- Andrew Grice
- Sean O’Grady
- Photography
- Theatre & Dance
- Culture Videos
- Fitness & Wellbeing
- Food & Drink
- Health & Families
- Royal Family
- Electric Vehicles
- Car Insurance Deals
- Lifestyle Videos
- UK Hotel Reviews
- News & Advice
- Simon Calder
- Australia & New Zealand
- South America
- C. America & Caribbean
- Middle East
- Politics Explained
- News Analysis
- Today’s Edition
- Home & Garden
- Broadband deals
- Fashion & Beauty
- Travel & Outdoors
- Sports & Fitness
- Sustainable Living
- Climate Videos
- Solar Panels
- Behind The Headlines
- On The Ground
- Decomplicated
- You Ask The Questions
- Binge Watch
- Travel Smart
- Watch on your TV
- Crosswords & Puzzles
- Most Commented
- Newsletters
- Ask Me Anything
- Virtual Events
- Betting Sites
- Online Casinos
- Wine Offers
Thank you for registering
Please refresh the page or navigate to another page on the site to be automatically logged in Please refresh your browser to be logged in
Melanie Road murder: How DNA collected in 1984 solved the 32-year-old case
Melanie road, 17, was found in a pool of blood at 5.30am on june 9 in 1984, article bookmarked.
Find your bookmarks in your Independent Premium section, under my profile

For free real time breaking news alerts sent straight to your inbox sign up to our breaking news emails
Sign up to our free breaking news emails, thanks for signing up to the breaking news email.
Christopher Hampton has been brought to justice after his daughter's DNA was put on the national database following a domestic incident with her partner.
Police first collected DNA samples from St Stephen's Court in Bath, Somerset, where Melanie Road, 17, was found in a pool of blood at 5.30am on June 9 in 1984.
She had been stabbed 26 times through her clothing, which Hampton removed to rape her and then placed back on her body.
A trail of blood matching Hampton's blood group, and later his DNA, was found leading from nearby St Stephen's Road in the direction of his home half a mile away.
- DNA probe over Brit backpacker pair murdered in Thailand 'incompetent'
- 'DNA factory' opens at Imperial College London
- Archbishop Justin Welby is illegitimate son of Churchill's secretary
- HIV-positive man guilty of kidnap and sexually assault of two girls
Officers from Avon and Somerset Police took swabs from 71 blood spots at the scene, as well as from Melanie's clothing and body.
DNA analysis was not available in 1984 but the force kept all the samples and evidence collected from the scene for more than three decades.
As scientific techniques advanced, various analyses were carried out and police persisted with case reviews.
A DNA profile of the killer, extracted from samples of semen from the scene, was loaded onto the national DNA database in the 1990s.
There was no match from the database at that time as Hampton had not been arrested for any criminal offences.
Scientific advances meant that familial DNA links could be identified, a breakthrough that proved crucial in solving Melanie's murder.
In 2014, Hampton's daughter, then aged 41, was arrested following a domestic incident with her partner in which a necklace was broken.
Police protocol for domestic violence meant she was cautioned and her DNA taken, then uploaded to the national database.
The following year, the familial DNA testing for Melanie's murder was re-run and a match was found with Hampton's daughter.
He voluntarily provided a mouth swab to retired police officer Paul Mason on June 1 in 2015, remaining calm and confident throughout.
This DNA was found to match samples taken from semen staining on the fly and crotch of Melanie's trousers.
Following his arrest, Hampton released a prepared statement saying: "I deny any involvement in the rape and murder of Melanie Road".
On December 22 last year, Hampton entered a not guilty plea to murder at Bristol Crown Court.
Scientific work continued and Hampton's DNA was found on four other areas of semen staining on Melanie's trousers and pants.
His DNA also matched semen on swabs taken from Melanie's body during a post-mortem examination.
It was also in blood spots found near Melanie's body.
Forensics: The Anatomy of Crime
Speaking outside court, Detective Chief Inspector Julie MacKay said: "The key to solving this case has been a combination of traditional police enquiries, advances in forensic science and the tenacity of a small group of officers and police staff.
"Although Hampton has now admitted to murdering Melanie, he has spent more than 30 years living a lie, able to conceal his dark secret from all those around him."
Ms MacKay told reporters she believed Hampton would have been eventually caught if it had not been for his daughter's DNA being put on the national database.
She praised the efforts of officers who worked on the 1984 investigation and said the senior investigating officer had died regretting Melanie's murder was unsolved.
"If in 1984 they hadn't swabbed every one of those 71 blood spots, if they hadn't meticulously examined everything that was available to them from Melanie's clothing and her body we would not have had that DNA available to us," she said.
Hampton was emotionless as the critical mouth swab was taken by Mr Mason.
"Bearing in mind he is a man who knew that he had taken that swab resulting in us identifying him I am amazed at how little emotion he showed," Mr Mason said.
"He was no different than any other swab and I have done hundreds. It was just another swab at that stage."
Subscribe to Independent Premium to bookmark this article
Want to bookmark your favourite articles and stories to read or reference later? Start your Independent Premium subscription today.
New to The Independent?
Or if you would prefer:
Want an ad-free experience?
Hi {{indy.fullName}}
- My Independent Premium
- Account details
- Help centre
The first exclusively online law review.
- Current Issue
- Symposium 2024
- Past Issues
- Scholastica
- Contributing to the JOLT Blog
- Admin Log In

Whose DNA is It Anyway? Legal Challenges that Arise from the Use of Genetic Genealogy in Criminal Investigations
On April 26, 2024
In Blog Posts
Since 2018, law enforcement’s use of genetic genealogy to identify and apprehend suspects has been growing [2] , especially in high profile cases like the Golden State Killer case and the recent University of Idaho student murders. However, it is not without its critics.
Jennifer Lynch, the general counsel of the non-profit civil liberties group Electronic Frontier Foundation, is one of these critics. In a recent interview with CNN , she cautions that “[genetic genealogy] is still a very new investigative technique” that is far from perfect. [3] In addition to possibly violating the 4 th Amendment against unlawful searches and seizures, Lynch points out that “the public should have the ability to know more about how these sorts of searches are conducted so that we can ensure the police are not just willy-nilly collecting DNA from people and running them against consumer genetic genealogy databases. She is hardly alone in her criticisms and concerns.
Genetic genealogy differs significantly from traditional criminal DNA searches [5] and the current state and federal laws are simply not enough to deal with the legal questions that are arising from its use. This post explores some of these concerns and explain why it is important for both federal and state government to act on this issue sooner rather than later.
Genetic genealogy is “a practice that blends DNA analysis in the lab with genealogical research, such as tracing a person’s family tree [6] .” The practice became popular in the last decade when commercial companies like 23andMe and Ancestry.com offered services where for a fee a customer could order a kit, submit a saliva sample for a DNA analysis, and then receive results that could tell a person about their ethnic and racial ancestry and how at risk they are for certain genetic health issues. Since these services first became available in 2007, millions of people have used them [7] . As of February 2024, it is estimated that one out of every five Americans has tried one of these services, and more than 14 million people alone have used 23andMe [8] .
While 23andMe does not share customer’s data with law enforcement, customers are still free to upload their raw DNA files from it and other similar services to sites with less protection like GEDMatch [9] . GEDMatch is a free, open-source site that was started in 2010 [10] . It takes the “raw DNA files that users upload and then compares them to the people who have already uploaded their name, email addresses and DNA data. [11] ” GEDMatch then analyses the results and produces a list of family relatives who have also opted in to its service [12] . The list of relatives includes everyone from parents to fourth and fifth cousins [13] . In addition, the list includes their contact information [14] . From there, genealogists take those relatives’ names and search through publicly available data bases for documents like birth certificates, obituaries and death certificates to build an even more extensive and detailed family tree. [15]
Although GEDMatch was originally intended for those seeking to learn more about their own family ancestry, law enforcement quickly realized that this could be a useful tool in helping to apprehend criminal suspects in cases with no known suspects but where DNA evidence was left behind at a crime scene [16] . The impact that GEDmatch has had on law enforcement cannot be understated. Since 2018, the information available through GEDMatch has led to an additional thirty-nine cold case arrests and played a role in identifying at least twelve previously unidentified remains [17] .
In the past, the protocol for when law enforcement found unknown DNA was to test it and then enter into the FBI-maintained CODIS DNA database where it was then compared to the DNA of known criminal offenders [18] . However, when there was no match, such as in the case of the aforementioned Golden State Killer case, suspects often remained unknown indefinitely.
By contrast, investigators can now take that unknown DNA and partner with genetics companies like Parabon NanoLabs to turn it into a raw data file [19] . That raw data file can then be upload to GEDMatch which in turn produces a list of extended family members who are related to the perpetrator [20] . That list can then allow investigators to narrow their search to on those who may have been in contact with the victim and are potential suspects [21] .
Usually, once the investigators have come up with that list of potential suspects, they then surveil them until they can obtain a discarded item that contains the suspect’s DNA which is then collected and tested to see if it matches the DNA found at the crime scene [22] . If it matches, then a warrant is sought to obtain a sample directly from the suspect [23] . If the sample confirms the earlier match from the discarded item, then police will make an arrest [24] . One of the biggest advantages to law enforcement is that these open databases “expands the number of possible suspects to include people who have not previously committed a crime. [25] ”
Expanding the number of possible suspects to include those who do not have criminal records is not the only way that genetic genealogy searches differ from traditional criminal DNA searches. DNA that is collected by law enforcement and entered into CODIS is “is subject to state and federal statutes that govern exactly how, when, and from whom it can be collected [26] .” DNA can only be collected from an arrest suspect who must be arrested under a warrant obtained on probable cause. That collected DNA must be expunged if a person is ultimately never charged or convicted [27] . Furthermore, “even the labs that process DNA entered into CODIS are subject to federal regulations and licensing requirements [28] .”
By contrast, genetic genealogy databases like GEDMatch are privately maintained. There are no regulations that regulate how these databases collect, handle, and share data [29] . They are also not covered under the Health Insurance Portability and Accountability Act (HIPAA) since HIPAA only applies to entities like healthcare providers and insurance companies [30] . There is no way of even guaranteeing that the DNA data files uploaded by users are actually their individual files other than the user’s word that is the case.
Another fundamental way in which traditional criminal DNA searches differ from genetic genealogy searches has to do with the overall quality of the samples that each typically uses to get their respective results. While private companies are able to control for quantity and contamination, most samples used in forensic application (especially those obtained from crime scenes) are often “degraded and contaminated samples [31] .”
While the recommended amount of DNA for testing is typically 200 nanograms of genetic material (ng), most forensic samples typically range between 0.1 ng to 1 ng [32] . As a result, using very small samples and multi-source samples for genetic genealogy searches may lead to serious errors or misidentifications [33] . While sibling matches may be able to withstand smaller samples, accuracy for second and third cousins falls significantly [34] . It is worth noting that in the cases of correctly identifying third cousins, even samples that were at the ideal of 200 ng were only accurate on average 74.6% of the time [35] . As a result, false positives can and do occur [36] . In addition to sample size, results are also affected by algorithmic parameters [37] . Therefore, without knowing the exact algorithms and parameters used to obtain the result, “it can be difficult to assess whether the results are reliable or are a product of manipulating bioinformatics [38] .”
The Fourth Amendment protects against unwanted searches and seizures [39] and is seen as the major source of privacy law in the United States [40] . While some originally interpreted it mostly as an effort to protect places, “the Supreme Court shifted Fourth Amendment doctrine from a property tort-based approach in Boyd v. United States to a reasonable expectations analysis in the famous wiretapping case Katz v. United States [41] . ” In Katz , Justice Harlan defined a new standard for judging reasonable search and seizure. “. . . [T]here is a twofold requirement, first that a person have exhibited an actual (subjective) expectation of privacy and, second, that the expectation be one that society is prepared to recognize as ‘reasonable [42] .”
The issue of what is reasonable is still a question of debate and is even more of a challenge in situations involving the use of genetic genealogy to apprehend suspects. The state successfully argued under California v. Greenwood , 486 U.S. 35 (1988) that the Fourth Amendment does not require a warrant to search DNA found on items that a suspect has discarded because the suspect abandoned their privacy interests in their DNA when they left it behind on those items. [43] Although Greenwood may not seem applicable in most cases involving the use of genetic genealogy,an argument could be made that when one decides to upload their DNA profile on a publicly shared data base they lose their right to DNA data privacy. It must be noted that in the vast majority of these recent cases including the Golden State Killer and the University of Idaho murders, the suspects did not actually upload their DNA themselves. Rather relatives, many of whom probably were not even aware of the existence of said suspects, were the ones who did that. Likewise, the third-party doctrine ruled that under the Fourth Amendment, an individual loses a reasonable expectation of when they volunteer information to a third party [44] would not appear to apply to these sort of suspects either since it is quite hard to argue that anyone “volunteers” their DNA data to a third party by virtue of being related to them.
Furthermore, there remains the question: What about the rights of those who did the actual uploading but never intended for their DNA data to be used for criminal procedures? Also, what about the commercial companies who wish to opt out as well? Examples abound of both users and commercial companies who specifically said that did not want DNA data to be used for the purposes of criminal investigation only to have those preferences ignored. Even a 2018 interim policy issued by the Justice Department on the use of familial genealogical genetics (FGG) by federal agencies that states investigative “shall identify themselves as law enforcement to services and enter and search FGG profiles only in those [genealogical genetic] services that provide explicit notice to their service users [45] ” failed to offer protection in some situations. For instance, it has been reported that a loophole on the GEDMatch website, which allowed researchers to view “the profiles of individuals who had specifically opted out of the use of their data in law enforcement sources.” [46] In summer 2023, it was revealed that California’s Riverside County Regional Cold Case Team uploaded a victim’s DNA to the commercial site MyHeritage despite the fact that doing so is a direct violation of the company’s term of services. It was further claimed that Riverside County Regional Cold Case Team may have acted with cooperation from the FBI. [47] Lastly, there is also the question of those individuals and companies that are comfortable with DNA data being used for investigative purpose in cases involving serious offensives like murder or sexual assaults but object to it being used in lesser crimes. As Electronic Frontier Foundation’s Jennifer Lynch said to CNN:
There’s a tendency for judges, for the public, to say, ‘Oh, well, these crimes are so terrible that it justifies any kind of search to possibly identify the person,’” she said. “But what we have to realize is that these kinds of investigative techniques are not limited to cold cases and not limited to heinous felonies. They will be used in even minor crimes, and they can implicate people for crimes they didn’t commit. [48]
Lynch’s statements about this being used for minor crimes are backed up by the data: In October 2019 it was announced that out of the ten requests made by law enforcement to access 23andMe’s data base, “[t]he majority of the requests concerned credit card fraud. [49]
As of this writing, there is no national standard for the use of genetic genealogy. Twelve states allow for the use of it while Montana and Maryland are the only two states that that have explicit statutory limitations on the use of genetic genealogy and familial searches [50] .
The lack of legal precedent on the subject is one of the many reasons why the University of Idaho murder case is drawing so much attention, not just from the usual true crime reporters but also legal reporters and scholars. While the judge has yet to set a date for the trial, it is likely the issue of the genetic genealogy will come up before, during, and possibly even after the trial.
The few existing state and federal laws that we currently have are not enough when it comes to the regulation of genetic genealogy, and its use in criminal investigation. Neither current laws nor past court rulings involving traditional DNA searches or reasonable expectations of privacy that were established prior to the advent of genetic genealogy technology are adequate to solve the challenges and questions that this new technology presents Answers to questions or even attempts to find answers to question like who exactly owns and controls our DNA-arguably our most personal of all property that we possess but are constantly shedding and leaving behind wherever we go and parts of which we share with others through no choice of our own-are not going to be easy tasks. However, we cannot wait for the courts and the states to slowly assemble a patchwork of laws. Instead, what is needed is both state and federal legislation that will protect 4 th Amendment rights even in a time where DNA which was once locked in mystery is increasingly becoming public information regardless of whether we provide consent or not.
Image Source: kjpargeter on Freeport.com
[1] Eric Levenson, It Started as a Hobby. Now They’re Using DNA to Help Cops Crack Cold Cases , CNN (Mar. 27, 2018, UPDATED 10:27 AM EDT), https://www.cnn.com/2018/08/03/health/dna-genealogy-cold-cases-trnd/index.html .
[3] Eric Levenson, A Hearing in the Idaho Student Killings Case Focuses on Genetic Genealogy. Here’s Why that May be Important , CNN (Feb. 29, 2024, UPDATED 9:45 AM EST),https://www.cnn.com/2024/02/28/us/idaho-student-killings-genetic-genealogy/index.html.
[5] Jennifer Lynch, Forensic Genetic Genealogy Searches: What Defense Attorneys & Policy Makers Need to Know , Electronic Frontier Foundation (Jul. 26, 2023), https://www.eff.org/wp/forensic-genetic-genealogy-searches-what-defense-attorneys-need-know#Regulation.
[6] Levenson, supra note 1.
[7] Alaina Demopoulos, ‘ There are no serious safeguards’: Can 23andMe be Trusted with our DNA?, The Guardian (Feb. 17, 2024, 12:00 PM EST), https://www.theguardian.com/technology/2024/feb/17/23andme-dna-data-securityfinance#:~:text=Of%20course%2C%20most%2023andMe%20experiences,learning%20more%20about%20their .
[9] Levenson, supra note 1.
[12] Levenson, supra note 1.
[16] Levenson, supra note 3.
[17] Genevieve Carter, The Genetic Panopticon: Genetic Genealogy Searches and the Fourth Amendment , 18 NW. J. TECH. & INTELL. PROP. 311, 321 (2021).
[18] Levenson, supra note 1.
[22] Levenson, supra note 1.
[26] Lynch, supra note 5.
[30] Genevieve Carter, The Genetic Panopticon: Genetic Genealogy Searches and the Fourth Amendment , 18 NW. J. TECH. & INTELL. PROP. 311, 317 (2021).
[31] Bicka Barlow & Kristen McCowan, Genetic Genealogy in the Legal System , 97 WIS. LAW. 14, 15 (2024).
[32] Id. at 16.
[33] Lynch, supra note 5.
[34] Barlow & McCowan, supra note 31, at 16.
[36] Lynch, supra note at 5.
[37] Barlow & McCowan, supra note 31, at 15.
[38] Id. , at 16.
[39] U.S. Cont. amend. IV.
[40] Carter, supra note 30 at 324.
[42] Katz v. United States , 389 U.S. 347, 88 S. Ct. 507, 588 (1967).
[43] Lynch , supra note 5.
[46] Bicka Barlow & Kristen McCowan, Genetic Genealogy in the Legal System , 97 WIS. LAW. 14, 17 (2024).
[48] Levenson, supra note 3.
[49] Genevieve Carter, The Genetic Panopticon: Genetic Genealogy Searches and the Fourth Amendment , 18 NW. J. TECH. & INTELL. PROP. 311, 317 (2021).
[50] Lynch, supra note 5.
Navigating Big Tech in Today’s Age of Antitrust Enforcement
Pluto: exploring robotics law through the lens of science fiction, leave a reply cancel reply.
You must be logged in to post a comment.
Powered by WordPress & Theme by Anders Norén

IMAGES
VIDEO
COMMENTS
NIJ's Solving Cold Cases with DNA Program. Since 2005, NIJ has awarded more than $73 million to more than 100 state and local law enforcement agencies through its Solving Cold Cases with DNA competitive grant program. This funding has allowed the agencies to review more than 119,000 cases. The funding has also facilitated the entry of almost ...
The next day brought another disturbing discovery: A county road worker found 16-year-old Patricia Kalitzke's body in an area north of Great Falls, the paper reports. She had been shot in the head ...
Lynette White was murdered in 1988. When the three men first imprisoned for her murder were found to have been wrongfully convicted, it seemed that her killer would go unpunished. However, new technology invented in 2002 was used to analyze DNA found at the scene of the murder. The only match was to a boy too young to have committed the murder ...
Now, DNA evidence points to a neighbor as her killer, police say ... Las Vegas police Lt. Ray Spencer announced Monday that advanced DNA testing helped investigators solve the cold case homicide ...
The first category of forensic DNA application can be illustrated with the Colin Pitchfork case in the 1980s, the first recorded case of DNA evidence used in a successful criminal investigation (BBC News, 2018). The case involved the rape and murder of two teenagers, Lynda Mann in 1983 and Dawn Ashworth in 1986.
NIJ funding helped the Boston Police Department solve a rape and murder case almost 50 years after the crime. was a ghastly crime. Nineteen-year-old Mary Sullivan had just moved from Cape Cod to Boston, where she rented an apartment in the bustling Beacon Hill neighborhood. Within a few days of her arrival in January 1964, she was found dead.
In the preceding chapters, we have tried to clarify the scientific issues involved in forensic DNA testing. This chapter discusses the legal implications of the committee's conclusions and recommendations. It describes the most important procedural and evidentiary rules that affect the use of forensic DNA evidence, identifies the questions of scientific fact that have been disputed in court ...
Case Study. Date Published: March 1, 2014. This article explains how the National Institute of Justice's (NIJ's) funding under its Solving Cold Cases with DNA program ("DNA program"), specifically its funded research on Y-STRs, was the key to solving the mystery of Mary Sullivan's rape and murder in Boston almost 50 years after her death.
Cold case DNA evidence is sometimes treated differently than in newer cases, says Pagliaro. "If the sample is degraded - affected by heat, UV light or moisture - the biologist may have to ...
We discuss herein some fascinating cases that involved the use of DNA as the primary evidence and have led to solving the mystery of these crimes. The first case study was of a case of sexual assault (Sect. 4.4.1). The second case study was of a case of sexual assault followed by the murder of an 8-year-old girl child.
Besides DNA evidence, hair and fiber examination also connected between Simpson and the crime scene. ... Dotson was released from the prison becoming the first person in history to be exonerated by the DNA evidence. Kirk Bloodsworth's Case. Kirk Noble Bloodsworth was sentenced to death in 1984 in a rape and murder case of a 9-year-old girl in ...
DNA typing has become the gold standard in forensic science since that first case 30 years ago. The advances in the 1970s paved the way for the detection of variation (polymorphism) in specific DNA sequences and shifted the study of human variation from the protein products of DNA to DNA itself (National Research Council (U.S.) 1992).As per the National Crime Records Bureau (NCRB) report ...
Hampikian conducted a similar study using DNA evidence from a real crime: the case of Kerry Robinson, a Georgia man serving 20 years for taking part in a gang rape. The victim had identified a man named Tyrone White as one of her attackers. Indeed, White's DNA matched 11 of the 13 alleles found in a DNA mixture at the crime scene that did not ...
Salting-Out Method. Introduced by Miller et al 55 in 1988, this method is a nontoxic DNA extraction method. Procedure: Sample is added to 3 mL of lysis buffer, SDS, and proteinase K, and incubated at 55 to 65°C overnight. Next, 6 mL of saturated NaCl is added and centrifuged at 2,500 rpm for 15 minutes.
The best evidence occurs when a person's DNA is found where it is not supposed to be. For example, consider a breaking-and-entering that occurred in a residential area. Near the point of forced entry, a knit cap was found which the homeowners confirm was not theirs. Several head hairs were recovered from the inside, one of which had a root ...
Killer breakthrough - the day DNA evidence first nailed a murderer. It's 30 years since DNA fingerprinting was first used in a police investigation. The technique has since put millions of ...
Forensic DNA analysis has vastly evolved since the first forensic samples were evaluated by restriction fragment length polymorphism (RFLP). Methodologies advanced from gel electrophoresis techniques to capillary electrophoresis and now to next generation sequencing (NGS). Capillary electrophoresis was and still is the standard method used in forensic analysis. However, dependent upon the ...
Officers from Avon and Somerset Police took swabs from 71 blood spots at the scene, as well as from Melanie's clothing and body. DNA analysis was not available in 1984 but the force kept all the ...
DNA Evidence and the Courts. Other sections of these materials highlight in more detail how improvements in sequencing DNA, storing DNA information, and interpreting those data are influencing the types of court cases that rely on DNA evidence. For example, there is growing interest in how improved DNA sequencing has led to the creation of ...
Whose DNA is It Anyway? Legal Challenges that Arise from the Use of Genetic Genealogy in Criminal Investigations. By Kim Lo. Since 2018, law enforcement's use of genetic genealogy to identify and apprehend suspects has been growing , especially in high profile cases like the Golden State Killer case and the recent University of Idaho student murders.